Biological data
Type of resources
Available actions
Topics
Keywords
Contact for the resource
Provided by
Years
Formats
Representation types
status
Resolution
-

GBIF, the Global Biodiversity Information Facility, is an international network and data infrastructure funded by the world's governments and aimed at providing anyone, anywhere, open access to data about all types of life on Earth. Coordinated through its Secretariat in Copenhagen, the GBIF network of participating countries and organizations, working through participant nodes, provides data-holding institutions around the world with common standards and open-source tools that enable them to share information about where and when species have been recorded. This knowledge derives from many sources, including everything from museum specimens collected in the 18th and 19th century to geotagged smartphone photos shared by amateur naturalists in recent days and weeks. The GBIF network draws all these sources together through the use of data standards, such as Darwin Core, which forms the basis for the bulk of GBIF.org's index of hundreds of millions of species occurrence records. Publishers provide open access to their datasets using machine-readable Creative Commons licence designations, allowing scientists, researchers and others to apply the data in hundreds of peer-reviewed publications and policy papers each year. Many of these analyses, which cover topics from the impacts of climate change and the spread of invasive and alien pests to priorities for conservation and protected areas, food security and human health, would not be possible without this. GBIF arose from a 1999 recommendation by the Biodiversity Informatics Subgroup of the Organization for Economic Cooperation and Development's Megascience Forum. This report concluded that "An international mechanism is needed to make biodiversity data and information accessible worldwide", arguing that this mechanism could produce many economic and social benefits and enable sustainable development by providing sound scientific evidence.
-

The GRSF, the Global Record of Stocks and Fisheries, integrates data from three authoritative sources: FIRMS (Fisheries and Resources Monitoring System), RAM (RAM Legacy Stock Assessment Database) and FishSource (Program of the Sustainable Fisheries Partnership). The GRSF content disseminated through this catalogue is distributed to test the logic to generate unique identifiers and review collated stock and fishery data. This beta release can contain errors and we welcome feedback on content and software performance, as well as the overall usability. Beta users are advised to use caution and to not rely in any way on the application and/or trust content and accompanying materials. What is the GRSF? A comprehensive and transparent inventory of stocks and fisheries records across multiple data providers.
-

OBIS is a global open-access data and information clearing-house on marine biodiversity for science, conservation and sustainable development. VISION: To be the most comprehensive gateway to the world’s ocean biodiversity and biogeographic data and information required to address pressing coastal and world ocean concerns. MISSION: To build and maintain a global alliance that collaborates with scientific communities to facilitate free and open access to, and application of, biodiversity and biogeographic data and information on marine life. More than 20 OBIS nodes around the world connect 500 institutions from 56 countries. Collectively, they have provided over 45 million observations of nearly 120 000 marine species, from Bacteria to Whales, from the surface to 10 900 meters depth, and from the Tropics to the Poles. The datasets are integrated so you can search and map them all seamlessly by species name, higher taxonomic level, geographic area, depth, time and environmental parameters. OBIS emanates from the Census of Marine Life (2000-2010) and was adopted as a project under IOC-UNESCO’s International Oceanographic Data and Information (IODE) programme in 2009. Objectives - Provide world’s largest scientific knowledge base on the diversity, distribution and abundance of all marine organisms in an integrated and standardized format (as a contribution to Aichi biodiversity target 19) - Facilitate the integration of biogeographic information with physical and chemical environmental data, to facilitate climate change studies - Contribute to a concerted global approach to marine biodiversity and ecosystem monitoring, through guidelines on standards and best practices, including globally agreed Essential Ocean Variables, observing plans, and indicators in collaboration with other IOC programs - Support the assessment of the state of marine biological diversity to better inform policymakers, and respond to the needs of regional and global processes such as the UN World Ocean Assessment (WOA) and the Intergovernmental Science-Policy Platform on Biodiversity and Ecosystem Services (IPBES) - Provide data, information and tools to support the identification of biologically important marine and coastal habitats for the development of marine spatial plans and other area-based management plans (e.g. for the identification of Ecologically or Biologically Significant marine Areas (EBSAs) under the Convention on Biological Diversity. - Increase the institutional and professional capacity in marine biodiversity and ecosystem data collection, management, analysis and reporting tools, as part of IOC’s Ocean Teacher Global Academy (OTGA) - Provide information and guidance on the use of biodiversity data for education and research and provide state of the art services to society including decision-makers - Provide a global platform for international collaboration between national and regional marine biodiversity and ecosystem monitoring programmes, enhancing Member States and global contributions to inter alia, the Global Ocean Observing System (GOOS) and the Global Earth Observing System of Systems (GEOSS)
-
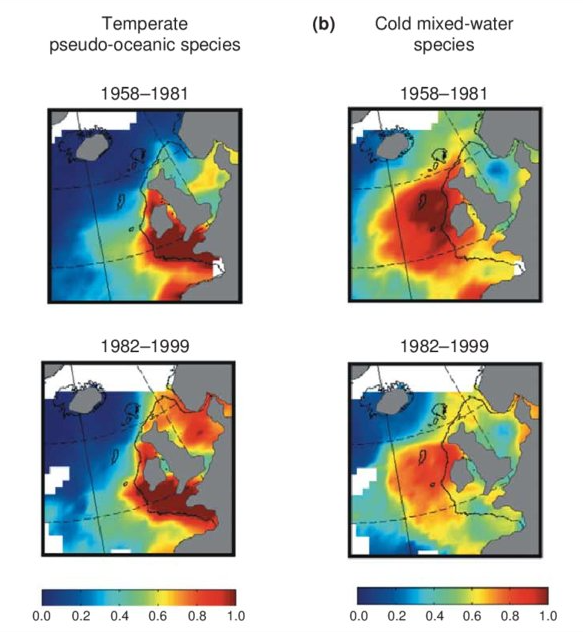
Excel file containing CPR data from Standard Areas B4,C3,C4,D3,D4,D5,E4,F4 for the plankton Calanus finmarchicus and helgolandicus, total traverse (small) copepods, total large copepods, Phytoplankton Colour Index and Cnidaria (presence denoted by a 1, absence by a zero). All taxa are from 1980, except Cnidaria which are from 2011. Dataset is in the format of sample level data, with each row being a discrete sample, with a sample being 3m3 filtered seawater, and 10nm of tow. For each row, a sample has the following information, starting at column a: Standard area of sample, sample id, latitude (decimal degrees) of sample mid point, longitude (decimal degrees) of sample midpoint, sample midpoint date and local time, year of sample, month of sample, then plankton abundance values (or PCI index or cnidaria presence/absence). All taxa have been looked for during the period this dataset spans, so zero values represent true absence.
-
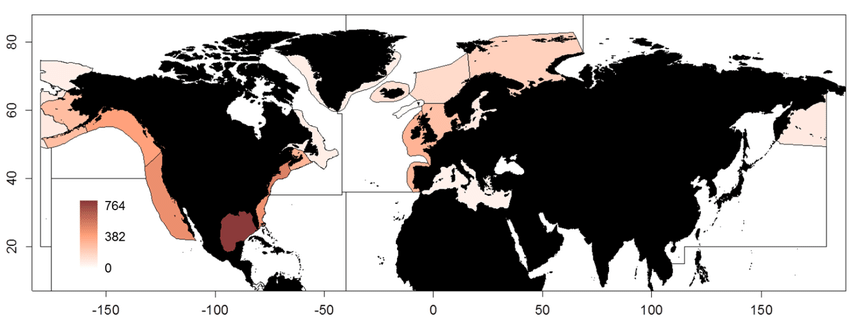
This dataset containing traits of marine fish is based on fish taxa observed during international scientific bottom-trawl surveys regularly conducted in the Northeast Atlantic, Northwest Atlantic and the Northeast Pacific. These scientific surveys target primarily demersal (bottom-dwelling) fish species, but pelagic species are also regularly recorded. The overarching aim of this dataset was to collect information on ecological traits for as many fish taxa as possible and to find area-specific trait values to account for intraspecific variation in traits, especially for widely distributed species. We collected traits for species, genera and families. The majority of trait values were sourced from FishBase (Froese and Pauly, 2019), and have been supplemented with values from the primary literature.
-
Accredited through the MEDIN partnership, and core-funded by the Department for the Environment, Food and Rural Affairs (Defra) and the Scottish Government, DASSH provides tools and services for the long-term curation, management and publication of marine species and habitats data, within the UK and internationally. Working closely with partners and data providers we are committed to the FAIR Data Principles, to make marine biodiversity data Findable, Accessible, Interoperable and Reusable. DASSH is a flagship initiative of the Marine Biological Association (MBA), and builds on the MBA's historic role in marine science. Through partnerships with other UK and European data centres DASSH contributes to data portals including the NBN Atlas, EMODnet, EurOBIS and GBIF. Fill out a form into the website to generate monthly lifeform abundances from the selected dataset.
-
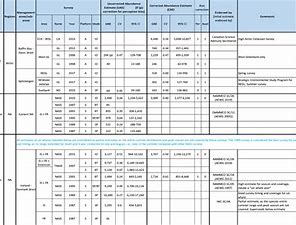
The purpose of this table is to present the best available abundance estimates for cetacean species in areas of relevance to the work of NAMMCO. It is intended to be used as a starting point for researchers, and the original sources are provided if additional information is required. The Scientific Committee of NAMMCO maintains a Working Group on Abundance Estimates, composed of invited experts in the field as well as some Committee members. This Working Group meets periodically to review new abundance estimates from recent surveys or, in some cases, re-analyses of older data. The reports of the Working Group are brought to the Scientific Committee at their annual meetings, and used to formulate advice on stock status, allowable removals or other matters. In most cases, the Scientific Committee will formally endorse estimates approved by the Working Group, and if so, this is indicated on the Table. Some estimates have been endorsed by the Scientific Committee of the International Whaling Commission (IWC).
-

The Plankton Lifeform Extraction Tool brings together disparate European plankton datasets into a central database from which it extracts abundance time series of plankton functional groups, called “lifeforms”, according to shared biological traits. This tool has been designed to make complex plankton datasets accessible and meaningful for policy, public interest, and scientific discovery. The Plankton Lifeform Extraction Tool currently integrates 155 000 samples, containing over 44 million plankton records, from nine different plankton datasets within UK and European seas, collected between 1924 and 2017. Additional datasets can be added, and time series can be updated.
-

SEAPOP (SEAbird POPulations) is a long-term monitoring and mapping programme for Norwegian seabirds that was established in 2005. The programme covers seabird populations in Norway, Svalbard and adjacent sea areas, and will provide and maintain base-line knowledge of seabirds for an improved management of this marine environment. The data analyses aim to develop further models of seabird distribution and population dynamics using different environmental parameters, and to explore the degree of covariation across different sites and species. This knowledge is urgently needed to distinguish human influences from those caused by natural variation.
-
.jpg)
NAMMCO is an international regional body for cooperation on conservation, management and study of cetaceans (whales, dolphins and porpoises) and pinnipeds (seals and walruses) in the North Atlantic. The members of NAMMCO — Faroe Islands, Greenland, Iceland and Norway — are committed to sustainable and responsible use of all living marine resources, including marine mammals. Through regional cooperation, the NAMMCO member countries aim to strengthen and further develop effective conservation and management measures for marine mammals. Acknowledging the rights and needs of coastal communities to make a sustainable living from what the sea can provide, such measures should be based on the best available scientific evidence and user knowledge and take into account the complexity and vulnerability of the marine ecosystem. The NAMMCO Agreement focuses on consolidating and advancing scientific knowledge of the North Atlantic marine ecosystem as a whole, and understanding better the role of marine mammals in this system. In 2017 NAMMCO member countries reaffirmed their cooperation through the Nuuk Declaration. NAMMCO Catch database: This database of reported catches is searchable and it is possible to filter the information by country, species or area. It is also possible to sort the different columns in ascending or descending order and to show up to 100 entries per page. Carry-over from previous years are included in the quota numbers, where applicable. NAMMCO aims to ensure that this database is kept up to date with correct and validated catch data. The reported catch data for harbour seals in Greenland prior to 2012 is not included in the database due to known sources of error. This includes identified errors in where harbour seal catch data has been entered in hunting reports, and cases of species misidentification.
 Catalogue PIGMA
Catalogue PIGMA