Crassostrea gigas hemocytes scRNA-seq sequences
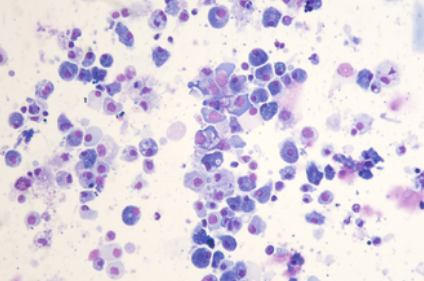
scRNA-seq reads from a Pacific oyster (Crassostrea gigas) hemocyte preparation. Hemocytes were isolated from a unique immunologically naive animal (Ifremer Standardized Animal, 18 months) and single-cell drop-seq technology was applied to 3,000 individual hemocytes.
Simple
- Date (Publication)
- 2024-10-31
- Date (Creation)
- 2020-02-13
- Identifier
- FR-330-715-368-00032-IFR_BIOINFO_CRASSOSTREA_GIGAS_20240209_IHPE_EV_HEMOFIGHT
- Credit
- IHPE UMR5244
Publisher
IFREMER
+33 (0)2 98.22.46.44
http://data.ifremer.fr/SISMER
IFREMER Centre de Bretagne ZI Pointe du diable CS 10070
,
PLOUZANE
,
29280
,
France
+33 (0)2 98.22.49.16
+33 (0)2 98.22.46.44
http://data.ifremer.fr/SISMER
Author
IFREMER (Institut Louis-Malardé, Univ Polynésie française)
-
Mitta Guillaume
Centre Océanologique du Pacifique - Vairao - BP 49 - 98179 Taravao - Tahiti - Polynésie Française
,
Tahiti
,
France
Author
IFREMER (IHPE UMR5244)
-
Courtay Gaelle
Université de Montpellier - Campus Triolet - 2 Place E. Bataillon - CC080 (IHPE) - 34095 Montpellier Cedex 5
,
Montpellier
,
France
04 67 14 47 10
- GEMET - INSPIRE themes, version 1.0
-
- Habitats and biotopes
- Thèmes Sextant
-
- /Biological Environment/Bioinformatics
- ODATIS aggregation parameters and Essential Variable names
-
- Bioinformatics
- Use limitation
- CC-BY (Creative Commons - Attribution)
- Access constraints
- Restricted
- Use constraints
- Copyright
- Other constraints
- Accès restreint jusqu'à la publication de l'article
- Metadata language
- English
- Character set
- UTF8
- Topic category
-
- Environment
N
S
E
W
))
- Reference system identifier
- EPSG / WGS 84 (EPSG:4326) / 8.6
- Distribution format
-
-
(
)
-
(
)
- OnLine resource
- /home/ref-bioinfo-public/ifremer/ihpe/hemofight ( NETWORK:LINK )
- OnLine resource
-
Lien de téléchargement
(
WWW:DOWNLOAD
)
Lien de téléchargement
- OnLine resource
- Publication ENA ( WWW:LINK )
- OnLine resource
- Digital Object Identifier (DOI) ( DOI )
- Hierarchy level
- Dataset
- Statement
- The 10X Genomics protocol was followed to prepare gel in emulsion beads containing single cells, hydrogel beads, and reverse transcription reagents, perform barcoded cDNA synthesis, and generate sequencing libraries from pooled cDNAs. The concentration of single cell suspension was approximately 1000 cells / μL, and cells were loaded according to the 10X protocol to capture approximately 3000 cells per reaction. Library construction was performed using 10X reagents according to the manufacturer. Libraries (paired end reads 75 bp) were sequenced on an Illumina NovaSeq using two sequencing lane per sample.
- File identifier
- ee5fd99f-d88f-4c6c-9b77-bfe1c88ca86f XML
- Metadata language
- English
- Character set
- UTF8
- Hierarchy level
- Dataset
- Date stamp
- 2024-07-22T15:22:06.703Z
- Metadata standard name
- ISO 19115:2003/19139 - SEXTANT
- Metadata standard version
- 1.0
Overviews
Spatial extent
N
S
E
W
))
Provided by

Associated resources
Not available
 Catalogue PIGMA
Catalogue PIGMA