2022
Type of resources
Available actions
Topics
Keywords
Contact for the resource
Provided by
Years
Formats
Representation types
Update frequencies
status
Service types
Scale
Resolution
-
French Zostera Marina et Zostera Noltei abundance data are collected during monitoring surveys on the English Channel / Bay of Biscay coasts. Protocols are impletmented in the Water Framework Directive. Data are transmitted in a Seadatanet format (CDI + ODV) to EMODnet Biology european database. 35 ODV files have been generated from period 01/01/2004 to 31/12/2021 for Z. Marina and from 01/01/2011 to 31/12/2021 for Z. Noltei.
-
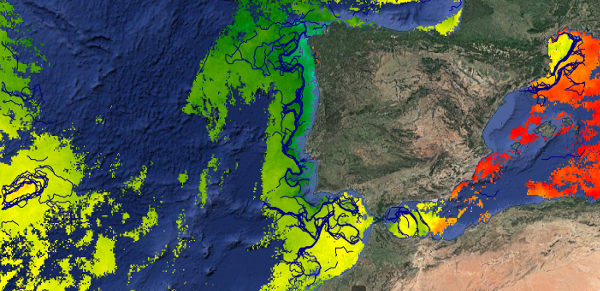
This dataset provides detections of fronts derived from high resolution remote sensing SST observations by SEVIRI L3C from OSISAF over Western Europe region. The data are available through HTTP and FTP; access to the data is free and open. In order to be informed about changes and to help us keep track of data usage, we encourage users to register at: https://forms.ifremer.fr/lops-siam/access-to-esa-world-ocean-circulation-project-data/ This dataset was generated by OceanDataLab and is distributed by Ifremer / CERSAT in the frame of the World Ocean Circulation (WOC) project funded by the European Space Agency (ESA).
-

In order to better characterize the genetic diversity of Cetaceans and especially the common Dolphin from the Bay of Biscay, sequences from the variable mitochondrial control region were obtained from water samples acquired close to groups of dolphins.
-

WGS for Iatlantic projet ( ) for assessing past and present connectivity
-
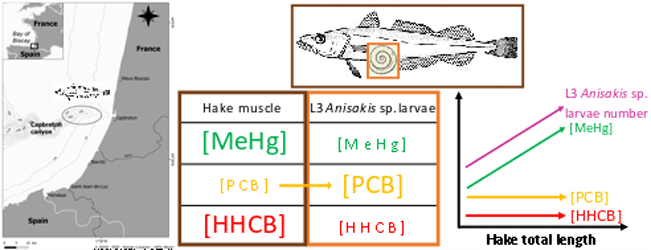
This dataset gathers isotopic ratios (carbon and nitrogen) and concentrations of both priority (mercury species and polychlorinated biphenyls congeners) and emerging (musks and sunscreens) micropollutants measured in a host-parasite couple (hake Merluccius merluccius muscle and in its parasite Anisakis sp) from the south of Bay of Biscay in 2018. In addition, the hake infection degree measured as the number of Anisakis sp. larvae was added for each hake collected.
-

This dataset consists of metatranscriptomic sequencing reads corresponding to coastal micro-eukaryote communities sampled in Western Europe in 2018 and 2019.
-
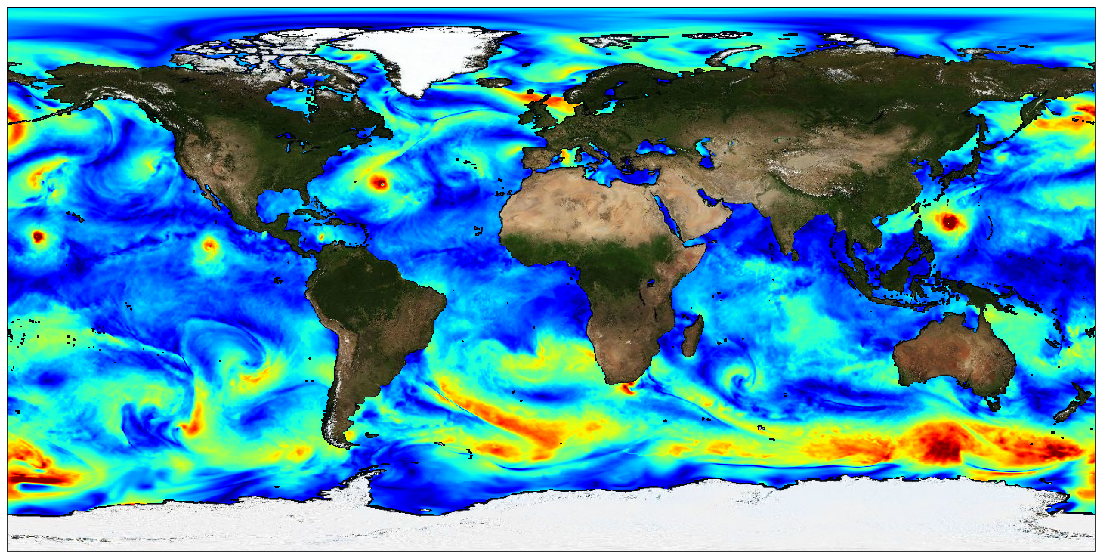
The Level 4 merged microwave wind product is a complete set of hourly global 10-m wind maps on a 0.25x0.25 degree latitude-longitude grid, spanning 1 Jan 2010 through the end of 2020. The product combines background neutral equivalent wind fields from ERA5, daily surface current fields from CMEMS, and stress equivalent winds obtained from several microwave passive and active sensors to produce hourly surface current relative stress equivalent wind analyses. The satellite winds include those from recently launched L-band passive sensors capable of measuring extreme winds in tropical cyclones, with little or no degradation from precipitation. All satellite winds used in the analyses have been recalibrated using a large set of collocated satellite-SFMR wind data in storm-centric coordinates. To maximize the use of the satellite microwave data, winds within a 24-hour window centered on the analysis time have been incorporated into each analysis. To accomodate the large time window, satellite wind speeds are transformed into deviations from ERA5 background wind speeds interpolated to the measurement times, and then an optical flow-based morphing technique is applied to these wind speed increments to propagate them from measurement to analysis time. These morphed wind speed increments are then added to the background wind speed at the analysis time to yield a set of total wind speeds fields for each sensor at the analysis time. These individual sensor wind speed fields are then combined with the background 10-m wind direction to yield vorticity and divergence fields for the individual sensor winds. From these, merged vorticity and divergence fields are computed as a weighted average of the individual vorticity and divergence fields. The final vector wind field is then obtained directly from these merged vorticity and divergence fields. Note that one consequence of producing the analyses in terms of vorticity and divergence is that there are no discontinuities in the wind speed fields at the (morphed) swath edges. There are two important points to be noted: the background ERA5 wind speed fields have been rescaled to be globally consistent with the recalibrated AMSR2 wind speeds. This rescaling involves a large increase in the ERA5 background winds beyond about 17 m/s. For example, an ERA5 10 m wind speed of 30 m/s is transformed into a wind speed of 41 m/s, and a wind speed of 34 m/s is transformed into a wind speed of about 48 m/s. Besides the current version of the product is calibrated for use within tropical cyclones and is not appropriate for use elsewhere. This dataset was produced in the frame of ESA MAXSS project. The primary objective of the ESA Marine Atmosphere eXtreme Satellite Synergy (MAXSS) project is to provide guidance and innovative methodologies to maximize the synergetic use of available Earth Observation data (satellite, in situ) to improve understanding about the multi-scale dynamical characteristics of extreme air-sea interaction.
-
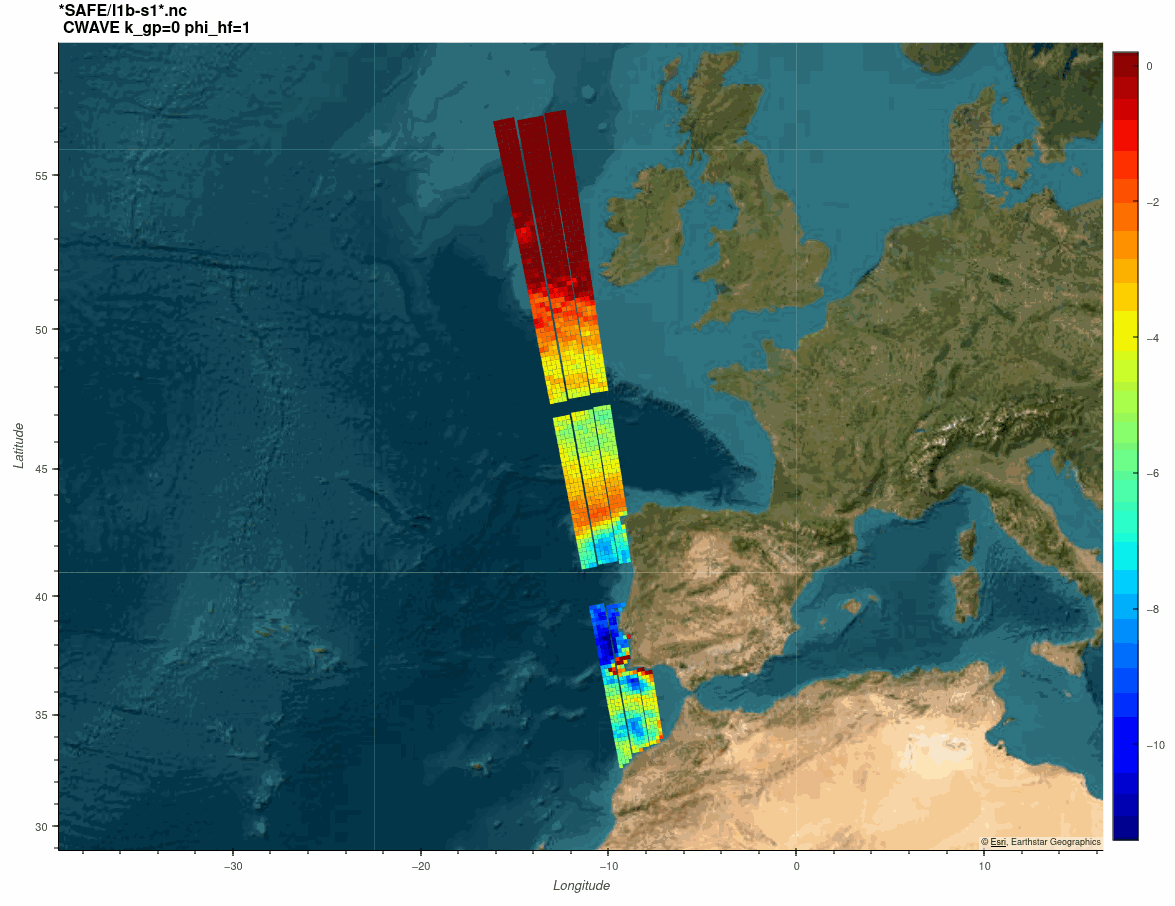
The SARWAVE project is developing a new sea state processor from SAR images to be applied over open ocean, sea ice, and coastal areas, and exploring potential synergy with other microwave and optical EO products.
-
The French Atlantic coast hosts numerous macrotidal and turbid estuaries that flow into the Bay of Biscay that are natural corridors for migratory fishes. The two best known are those of the Gironde and the Loire. However, there are also a dozen estuaries set geographically among them, of a smaller scale. The physico-chemical quality of estuarine waters is a necessary support element for biological life and determines the distribution of species, on which many ecosystem services (e.g. professional or recreational fishing) depend. With rising temperatures and water levels, declining precipitation and population growth projected for the New Aquitaine region by 2030, the question of how the quality and ecological status of estuarine waters will evolve becomes increasingly critical. The MAGEST (Mesures Automatisées pour l’observation et la Gestion des ESTuaires nord aquitains) high-frequency monitoring of key physico-chemical parameters was first developed in the Gironde estuary in 2004 ; the Seudre and Charente estuaries were instrumented late 2020. First based on real-time automated systems, MAGEST is now equipped by autonomous multiparameter sensors. Depending of the stations, an optode is also deployed to secure dissolved oxygen measurement. By the end of 2020, MAGEST had 12 instrumented sites. Portets is a measuring station located in the upper Gironde estuary (Garonne subestuary, about 20 km upstream of the Bordeaux metropolis.
-
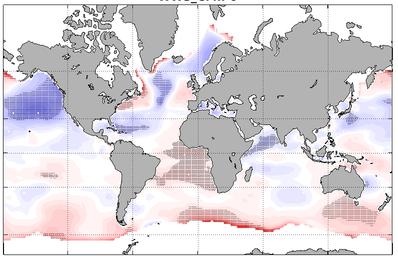
Global wave hindcast (1961-2020) at 1° resolution using CMIP6 wind and sea-ice forcings for ALL (historical), GHG (historical greenhouse-gas-only), AER (historical Anthropogenic-aerosol-only), NAT (historical natural only) scenario.
 Catalogue PIGMA
Catalogue PIGMA