*
Type of resources
Available actions
Topics
Keywords
Contact for the resource
Provided by
Years
Formats
Representation types
Update frequencies
status
Service types
Scale
Resolution
-
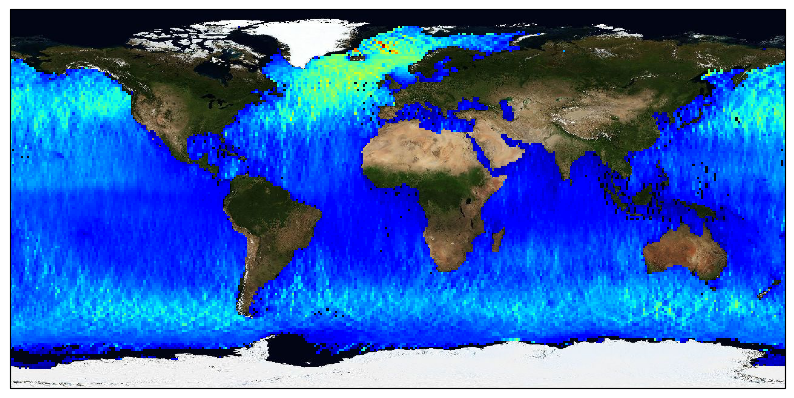
The ESA Sea State Climate Change Initiative (CCI) project has produced global multi-sensor time-series of along-track satellite altimeter significant wave height data (referred to as Level 4 (L4) data) with a particular focus for use in climate studies. This dataset contains the Version 3 Remote Sensing Significant Wave Height product, gridded over a global regular cylindrical projection (1°x1° resolution), averaging valid and good measurements from all available altimeters on a monthly basis (using the L2P products also available). These L4 products are meant for statistics and visualization. The altimeter data used in the Sea State CCI dataset v3 come from multiple satellite missions spanning from 2002 to 2021 ( Envisat, CryoSat-2, Jason-1, Jason-2, Jason-3, SARAL, Sentinel-3A), therefore spanning over a shorter time range than version 1.1. Unlike version 1.1, this version 3 involved a complete and consistent retracking of all the included altimeters. Many altimeters are bi-frequency (Ku-C or Ku-S) and only measurements in Ku band were used, for consistency reasons, being available on each altimeter but SARAL (Ka band).
-
-

-
Auteur(s): Baguet Hélène , Présentation des salles de spectacle à Bordeaux, puis de quelques salles en France. Le concept de Zénith est défini par une salle de grande capacité (3.000 places minimum) et un mode de gestion par une société privée laquelle offre un outil aux professionnels du spectacle : 13 bâtiments existants ou en cours de construction sont analysés. Projet d'un Zénith à Bordeaux, sur la friche Lauzun, dans le quartier du lac.
-

-
Map of seasonal averages of dissolved inorganic Nitrogen (uM) indicator for eutrophication for the past 10 years (2005-2014) in the Atlantic basin. It will be generated using in situ measurements of the different parameteres required to assess the dissolved inorganic Nitrogen indicator and the OSPAR Convention Common procedure methodology (OSPAR 2013, Common Procedure for the Identification of the Eutrophication Status of the OSPAR Maritime Area. Agreement 2013-08. 67 pp).
-
Auteur(s): Brun Jean-Pierre, Castagne Michel , La mer matrice originelle de toute vie, est encore présente lorsque l'homme ressent la nécessité de soins pour une remise en forme. L'aspect technique de la plate-forme, sa forme géométrique, le choix des matériaux, l'architecture high tech de la structure en tubes et en câbles qui soulagent la structure tridimensionnelle de la toiture, tous ces éléments aboutissent à une architecture géométrique, presque industrielle. Lorsque le curiste arrivera dans le centre, après avoir franchi la passerelle, il pénétrera dans un monde différent, fait de formes douces évoquant la fluidité de l'eau, image de ce retour aux sources de la vie
-
Temporal series (annual mean values) with error of estimation and Long Term Average (LTA) with error of estimation of total phosphate load for each river mouth where in situ data is available. Different sources can be mixed if any.
-

Description of attributes for time series of annual average sea level (units: mm) from tide gauges over periods of 50 years (1963-2012) and 100 years (1913-2012), to characterize and assess average annual sea-level rise relative to the land.
-
 Catalogue PIGMA
Catalogue PIGMA