Bioinformatics
Type of resources
Available actions
Topics
Keywords
Contact for the resource
Provided by
Years
Representation types
-

WGS of SARS-CoV-2 by Oxford Nanopore Technology from raw wastewater samples collected in France, 2020-2021
-

Individuals from 5 populations were kept in common garden conditions in order to examine acclimation and adaptation to temperature in the honeycomb worm. Worms were exposed to 5 temperature treatments, and collected for RNAseq analysis. Gene expression patterns were then examined.
-

Successive infections with Vibrio harveyi were conducted in two populations of the European abalone in order to examine which genes may be involved in improved survival to the disease in the St. Malo population.
-

This dataset provides an overview of the diversity of bacterial communities present in faeces and effluents of different origins and particularly in the droppings of wild birds in France. The aim is to develop new molecular markers associated with these different origins/hosts to identify the origin of faecal contamination in the environment.
-
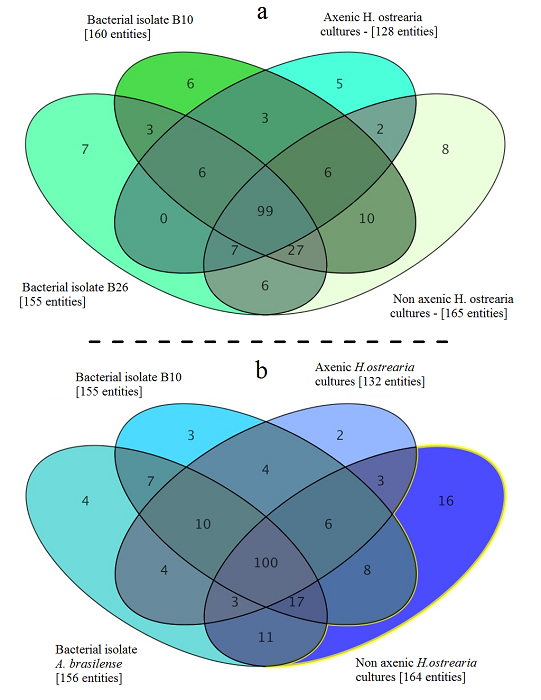
Metabolome of of the marine diatom Haslea ostrearia. Bacteria were isolated from Haslea ostrearia isolates cultivated in ES 1/3 medium in laboratory conditions over a 3-month period. These microalgal isolates were recovered from four sites on the French Atlantic coast: Bouin , La Barre-de-Monts (46.90 N; 2.11°W), Isle de Ré (46.22 N; 1.45°W), and La Tremblade (45.80 N; 1.15°W) . Data processing and statistical analysis of the metabolic profiles were performed on an LC/MS Metabolomics Discovery Workflow using Mass Profiler Professional Software and an Agilent 1290 Infinity II LC system coupled to an Agilent 6540 UHD Accurate-Mass QTOF hybrid mass spectrometer (Agilent Technologies, Waldbronn, Germany) equipped with a dual electrospray ionization (ESI) source. The full history (tools, parameters, input and output data files) is publicly available on http://dx.doi.org/10.12770/046e1e6a-864e-48a6-944b-d8613d67de0f
-

DNA sequencing of Crassostrea gigas Pacific oyster spat infected in the wild with OsHV-1 virus in 4 French oyster basins (Marennes Oleron Bay, Arcachon bay, Rade de Brest and Thau lagoon).
-
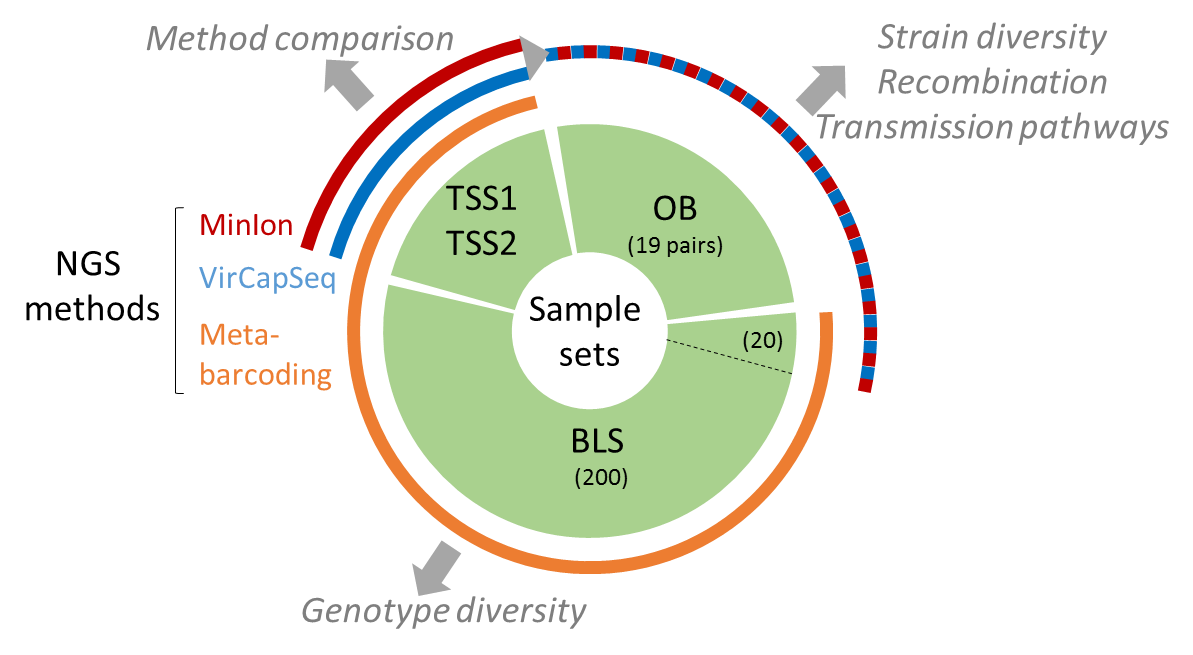
The present data set concerne metabarcoding raw reads produced using 4 different PCR targeting polymerase or capside coding region of the genoyupe I and II of norovirus. Test samples of norovirus with serial dilutions in pure water and after a bio-accumulation in oysters. Sequencing was made after VirCapSeq-VERT approach.
-

ddRAD genotyping was used to evaluate population connectivity and putative loci under selection in honeycomb worm from 13 sites spanning its distribution in the Atlantic and Mediterranean coasts.
-

WGS for Iatlantic projet ( ) for assessing past and present connectivity
-
 Catalogue PIGMA
Catalogue PIGMA