2016
Type of resources
Available actions
Topics
Keywords
Contact for the resource
Provided by
Years
Formats
Representation types
Update frequencies
status
Scale
Resolution
-

Description of attributes for time series of sea level trend for the last 10 yrs for the Mediterranean basin and for each NUTS3 region along the coast.
-
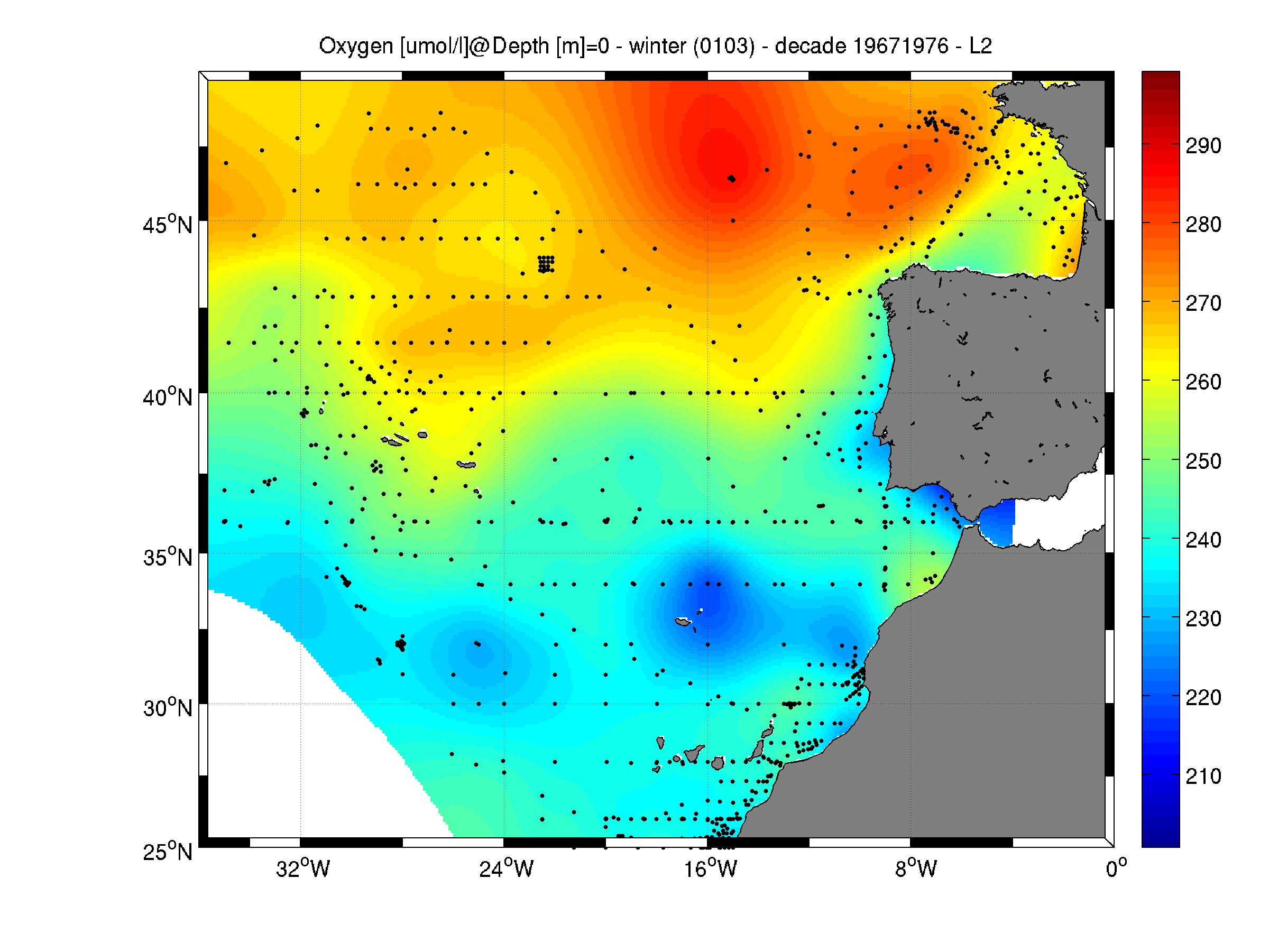
Moving 10-years analysis of Oxygen at Northeast Atlantic Ocean for each season: - winter: January-March, - spring: April-June, - summer: July-September, - autumn: October-December. Every year of the time dimension corresponds to the 10-year centred average of each season. Decades span from 1963-1972 until 2005-2014. Observational data span from 1963 to 2014. Depth range (IODE standard depths): -3000.0, -2500.0, -2000.0, -1750, -1500.0, -1400.0, -1300.0, -1200.0, -1100.0, -1000.0, -900.0, -800.0, -700.0, -600.0, -500.0, -400.0, -300.0, -250.0, -200.0, -150.0, -125.0, -100.0, -75.0, -50.0,-40.0, -30.0, -20.0, -10.0, -5.0, -0.0 Data Sources: observational data from SeaDataNet/EMODNet Chemistry Data Network. Description of DIVA analysis: Geostatistical data analysis by DIVA (Data-Interpolating Variational Analysis) tool. GEBCO 1min topography is used for the contouring preparation. Analyzed filed masked using relative error threshold 0.3 and 0.5 DIVA settings. Signal to noise ratio and correlation length were optimized and filtered vertically and a seasonally-averaged profile was used. Background field: the data mean value is subtracted from the data. Detrending of data: no, Advection consraint applied: no. Units: umol/l
-

Moving 10-years analysis of Phosphate at Northeast Atlantic Ocean for each season: - winter: January-March, - spring: April-June, - summer: July-September, - autumn: October-December Every year of the time dimension corresponds to the 10-year centred average of each season. Decades span : - from 1963-1972 until 2005-2014 (winter) - from 1963-1972 until 2005-2014 (spring) - from 1964-1973 until 2005-2014 (summer) - from 1964-1973 until 2005-2014 (autumn) Observational data span from 1962 to 2014. Depth range (IODE standard depths): -3000.0, -2500.0, -2000.0, -1750, -1500.0, -1400.0, -1300.0, -1200.0, -1100.0, -1000.0, -900.0, -800.0, -700.0, -600.0, -500.0, -400.0, -300.0, -250.0, -200.0, -150.0, -125.0, -100.0, -75.0, -50.0,-40.0, -30.0, -20.0, -10.0, -5.0, -0.0 Data Sources: observational data from SeaDataNet/EMODNet Chemistry Data Network. Description of DIVA analysis: Geostatistical data analysis by DIVA (Data-Interpolating Variational Analysis) tool. GEBCO 1min topography is used for the contouring preparation. Analyzed filed masked using relative error threshold 0.3 and 0.5 DIVA settings. Signal to noise ratio and correlation length were optimized and filtered vertically and a seasonally-averaged profile was used. Logarithmic transformation applied to the data prior to the analysis. Background field: the data mean value is subtracted from the data. Detrending of data: no, Advection constraint applied: no. Units: umol/l
-

The objective of this tender is to examine the current data collection, observation and data assembly programmes in the Meditterranean Sea, identify gaps and to evaluate how they can be optimised.
-

Description of the attributes for the time-series annual average sea level for the last 50 and the last 100 yrs for the Mediterranean basin and for each NUTS region along the coast.
-
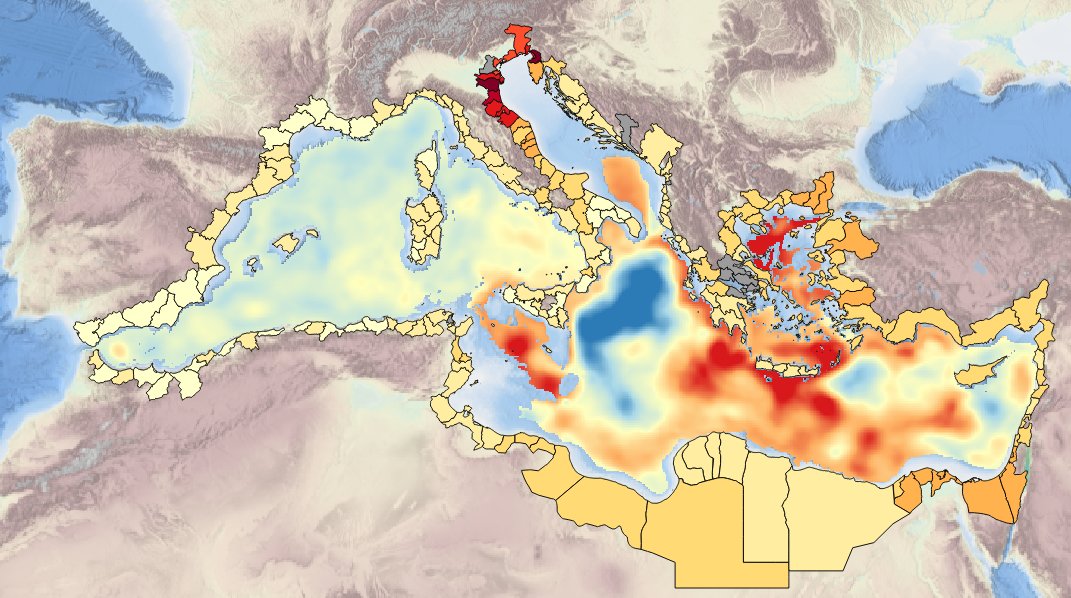
Specification of the desirable and recommended products attributes for generating spatial layers of sea mid-water and sea-bottom temperature for the last 10, 50 and 100 years for the Mediterranean basin and for each NUTS3 region along the coast.
-
Data from FerryBoxes on ships of opportunity going on permanent routes are stored inside this database (ferrydata.hzg.de). Parameters are temperature, salinity, chlorophyll-a fluorescence, oxygen and different others. The data model is transect oriented. A data portal to access and visualise the data is also provided.
-
'''This product has been archived''' For operationnal and online products, please visit https://marine.copernicus.eu '''Short description''' The Operational Mercator global ocean analysis and forecast system at 1/12 degree is providing 10 days of 3D global ocean forecasts updated daily. The time series is aggregated in time in order to reach a two full year’s time series sliding window. This product includes daily and monthly mean files of temperature, salinity, currents, sea level, mixed layer depth and ice parameters from the top to the bottom over the global ocean. It also includes hourly mean surface fields for sea level height, temperature and currents. The global ocean output files are displayed with a 1/12 degree horizontal resolution with regular longitude/latitude equirectangular projection. 50 vertical levels are ranging from 0 to 5500 meters. This product also delivers a special dataset for surface current which also includes wave and tidal drift called SMOC (Surface merged Ocean Current). '''DOI (product) :''' https://doi.org/10.48670/moi-00016
-
Confidence in the full output of the 2016 EUSeaMap broad-scale predictive model, produced by EMODnet Seabed Habitats. Values are on a range from 1 (low confidence) to 3 (high confidence). Confidence is calculated by amalgamating the confidence values of the underlying applicable habitat descriptors used to generate the habitat value in the area in question. Habitat descriptors differ per region but include: Biological zone Energy class Oxygen regime Salinity regime Seabed Substrate Riverine input Confidence in habitat descriptors are driven by the confidence in the source data used to determine the descriptor, and the confidence in the threshold/margin (areas closer to a boundary between two classes will have lower confidence). Confidence values are also available for each habitat descriptor and input data layer.
-
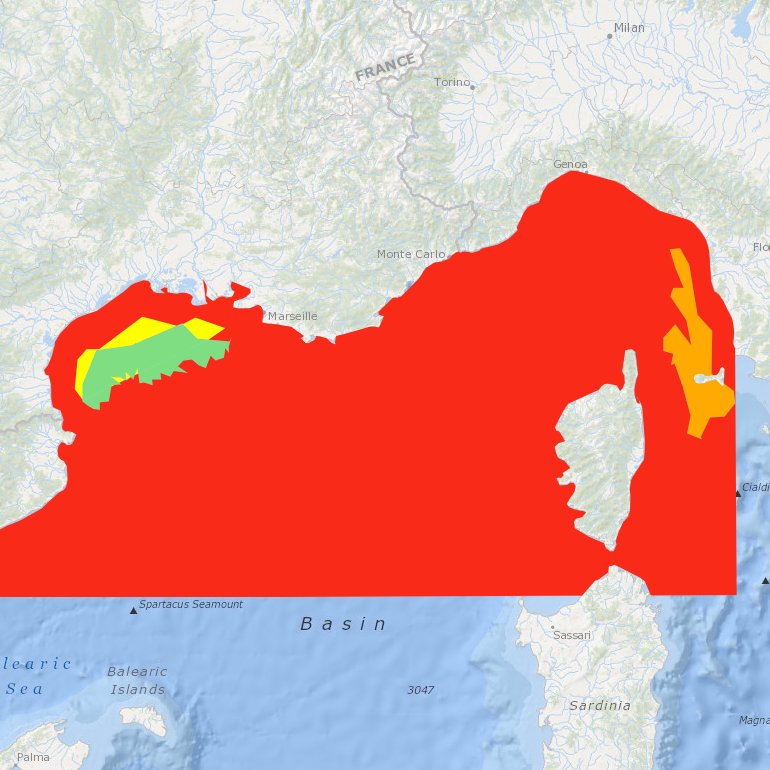
Suitability index of a wind farm in the NWMed concerning the environmental resources, the natural barriers, human activities, MPA and fisheries.
 Catalogue PIGMA
Catalogue PIGMA