2022
Type of resources
Available actions
Topics
Keywords
Contact for the resource
Provided by
Years
Formats
Representation types
Update frequencies
status
Service types
Scale
Resolution
-
The ESA Sea State Climate Change Initiative (CCI) project has produced global multi-sensor time-series of along-track satellite synthetic aperture radar (SAR) integrated sea state parameters (ISSP) data from ENVISAT (referred to as SAR Wave Mode onboard ENVISAT Level 2P (L2P) ISSP data) with a particular focus for use in climate studies. This dataset contains the ENVISAT Remote Sensing Integrated Sea State Parameter product (version 1.1), which forms part of the ESA Sea State CCI version 3.0 release. This product provides along-track significant wave height (SWH) measurements at 5km resolution every 100km, processed using the Li et al., 2020 empirical model, separated per satellite and pass, including all measurements with flags and uncertainty estimates. These are expert products with rich content and no data loss. The SAR Wave Mode data used in the Sea State CCI SAR WV onboard ENVISAT Level 2P (L2P) ISSP v3 dataset come from the ENVISAT satellite mission spanning from 2002 to 2012.
-

The CDR-derived Wet Tropospheric Correction (WTC) Product V2 is generated from the Level-2+ along-track altimetry products version 2024 (L2P 2024) distributed by AVISO+ (www.aviso.altimetry.fr). It provides a long-term, homogenized estimation of the wet tropospheric correction based on Climate Data Records (CDRs) of atmospheric water vapour combined with high frequencies MWR data. Two independent CDRs datasets are used: - REMSS V7R2 (coverage until 2022) https://www.remss.com/measurements/atmospheric-water-vapor/tpw-1-deg-product/ - HOAPS V5 precursor CDR from EUMETSAT CM SAF (coverage until 2020) HOAPS V4/V5 data available via https://wui.cmsaf.eu Note: the HOAPS V5 precursor is not yet an official CM SAF product; full validation and public release are pending. The MWR/CDR WTC V2 estimates is derived using spatially varying but temporally constant polynomial coefficients (ai). 1. WTC V2 – Along-track L2P Product Data format: The WTC V2 product is delivered in Level-2+ (L2P) format, along the satellite ground track. Each mission is distributed as a compressed archive (.tar.gz) containing one NetCDF4 CF-1.8 file per mission cycle. Archive naming convention: <mission>_WTC_from_WV_CDR_<version>.tar.gz mission: TP (TOPEX/Poseidon), J1, J2, J3 version: product version (currently V2) File naming convention inside archives: <mission>_C<cycle>.nc cycle: 4-digit cycle index (e.g., C0001) Each NetCDF file contains: 1/ Along-track WTC estimate; 2/ Ancillary information; 3/ Space–time coordinates 2. WTC CDR Uncertainties – Gridded Product: A complementary product is provided, delivering regional trend estimates and associated uncertainties from the WTC Climate Data Record. The uncertainty product is distributed as a single NetCDF4 file: wtc_trend_uncertainties.nc . This file contains global gridded fields of WTC CDR trend and uncertainty parameters. Product content: This is the first dedicated version providing both: WTC CDR (HOAPS) linear trends, and Uncertainty estimates on these trends. Uncertainties are expressed as 1-sigma confidence intervals, and propagated using the methodology described in Section 2.3 of the Product User Manual. The product includes: - Total uncertainty on the WTC trend, propagated from all identified uncertainty sources in the WTC–TCWV regression. - Individual contributions of uncertainty sources (Uncertainties on regression coefficients: a0, a1 and their standard deviations; Uncertainties inherited from the HOAPS TCWV CDR) These fields enable users to assess the relative importance of each uncertainty component and recompute uncertainty propagation with alternative methods. Included regression input variables: To ensure transparency and reproducibility, the product provides: 1/ regression coefficients a0, a1; 2/ their associated uncertainties (std of a0, std of a1); 3/additional diagnostic fields required to recompute uncertainties if needed.
-
French Zostera Marina et Zostera Noltei abundance data are collected during monitoring surveys on the English Channel / Bay of Biscay coasts. Protocols are impletmented in the Water Framework Directive. Data are transmitted in a Seadatanet format (CDI + ODV) to EMODnet Biology european database. 35 ODV files have been generated from period 01/01/2004 to 31/12/2021 for Z. Marina and from 01/01/2011 to 31/12/2021 for Z. Noltei.
-
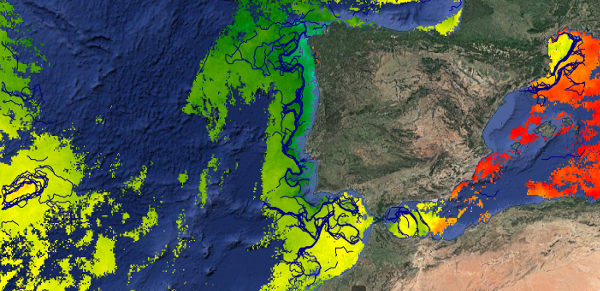
This dataset provides detections of fronts derived from high resolution remote sensing SST observations by SEVIRI L3C from OSISAF over Western Europe region. The data are available through HTTP and FTP; access to the data is free and open. In order to be informed about changes and to help us keep track of data usage, we encourage users to register at: https://forms.ifremer.fr/lops-siam/access-to-esa-world-ocean-circulation-project-data/ This dataset was generated by OceanDataLab and is distributed by Ifremer / CERSAT in the frame of the World Ocean Circulation (WOC) project funded by the European Space Agency (ESA).
-
Serveur wms sur les photos anciennes
-
Serveur wms du projet CHARM II
-

In order to better characterize the genetic diversity of Cetaceans and especially the common Dolphin from the Bay of Biscay, sequences from the variable mitochondrial control region were obtained from water samples acquired close to groups of dolphins.
-

WGS for Iatlantic projet ( ) for assessing past and present connectivity
-
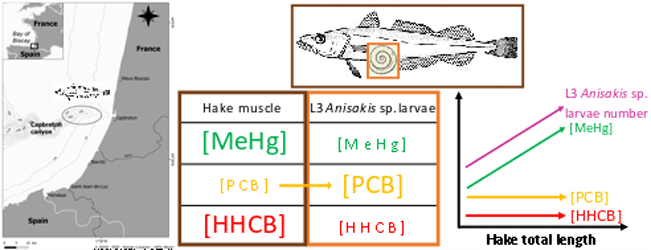
This dataset gathers isotopic ratios (carbon and nitrogen) and concentrations of both priority (mercury species and polychlorinated biphenyls congeners) and emerging (musks and sunscreens) micropollutants measured in a host-parasite couple (hake Merluccius merluccius muscle and in its parasite Anisakis sp) from the south of Bay of Biscay in 2018. In addition, the hake infection degree measured as the number of Anisakis sp. larvae was added for each hake collected.
-

Phenotypic plasticity, the ability of a single genotype to produce multiple phenotypes, is important for survival when species are faced with novel conditions. Theory predicts that range-edge populations will have greater phenotypic plasticity than core populations, but empirical examples from the wild are rare. The honeycomb worm, Sabellaria alveolata (L.), constructs the largest biogenic reefs in Europe, which support high biodiversity and numerous ecological functions. In order to assess the presence, causes and consequences of intraspecific variation in developmental plasticity and thermal adaptation in the honeycomb worm, we carried out common-garden experiments using the larvae of individuals sampled from along a latitudinal gradient covering the entire range of the species. We exposed larvae to three temperature treatments and measured phenotypic traits throughout development. We found phenotypic plasticity in larval growth rate but local adaptation in terms of larval period. The northern and southern range-edge populations of S. alveolata showed phenotypic plasticity for growth rate: growth rate increased as temperature treatment increased. In contrast, the core range populations showed no evidence of phenotypic plasticity. We present a rare case of range-edge plasticity at both the northern and southern range limit of species, likely caused by evolution of phenotypic plasticity during range expansion and its maintenance in highly heterogeneous environments. This dataset presents the raw image data collected for larval stages of Sabellaria alveolata from 5 populations across Europe and Northern Africa, exposed to 15, 20 and 25 C. Included are also opercular crown measurements used to estimate de size classes of individuals present in each population. All measurements made with the images collected are presented in an Excel spreadsheet, also available here.
 Catalogue PIGMA
Catalogue PIGMA