ZooScan
Type of resources
Available actions
Topics
Keywords
Contact for the resource
Provided by
Years
Formats
-

Plankton was sampled with various nets, from bottom or 500m depth to the surface, in many oceans of the world. Samples were imaged with a ZooScan. The full images were processed with ZooProcess which generated regions of interest (ROIs) around each individual object and a set of associated features measured on the object (see Gorsky et al 2010 for more information). The same objects were re-processed to compute features with the scikit-image toolbox http://scikit-image.org. The 1,451,745 resulting objects were sorted by a limited number of operators, following a common taxonomic guide, into 98 taxa, using the web application EcoTaxa http://ecotaxa.obs-vlfr.fr. For the purpose of training machine learning classifiers, the images in each class were split into training, validation, and test sets, with proportions 70%, 15% and 15%. taxa.csv.gz Table of the classification of each object in the dataset, with columns : - objid: unique object identifier in EcoTaxa (integer number). - taxon_level1: taxonomic name corresponding to the level 1 classification - lineage_level1: taxonomic lineage corresponding to the level 1 classification - taxon_level2: name of the taxon corresponding to the level 2 classification - plankton: if the object is a plankton or not (boolean) - set: class of the image corresponding to the taxon (train : training, val : validation, or test) - img_path: local path of the image corresponding to the taxon (of level 1), named according to the object id features_native.csv.gz - objid: same as above - area: area - mean: mean grey - stddev: standard deviation of greys - mode: modal grey - min: minimum grey - max: maximum grey - perim.: perimeter - width,height dimensions - major,minor: length of major,minor axis of the best fitting ellipse - circ.: circularity: 4pi(area/perim.^2) - feret: maximal feret diameter - intden: integrated density: mean*area - median: median grey - skew,kurt: skewness,kurtosis of the histogram of greys - %area: proportion of the image corresponding to the object - area_exc: area excluding holes - fractal: fractal dimension of the perimeter - skelarea: area of the one-pixel wide skeleton of the image - slope: slope of the cumulated histogram of greys - histcum1,2,3: grey level at quantiles 0.25, 0.5, 0.75 of the histogram of greys - nb1,2,3: number of objects after thresholding at the grey levels above - symetrieh,symetriev: index of horizontal,vertical symmetry - symetriehc,symetrievc: same but after thresholding at level histcum1 - convperim,convarea: perimeter,area of the convex hull of the object - fcons: contrast - thickr: thickness ratio: maximum thickness/mean thickness - esd: Equivalent Spherical Diameter - elongation: elongation index: major/minor - range: range of greys: max-min - centroids : sqrt(pow(xm-x,2)+ pow(ym-y,2)) - sr: index of variation of greys: 100*(stddev/range) - perimareaexc : perim/(sqrt(area_exc)) - feretareaexc : feret/(sqrt(area_exc)) - perimferet: index of the relative complexity of the perimeter: perim/feret - perimmajor: index of the relative complexity of the perimeter: perim/major - circex: (4*PI*area_exc)/(pow(perim,2)) - cdexc: (1/(sqrt(area_exc))) * sqrt(pow(xm-x,2)+pow(ym-y,2) features_skimage.csv.gz Table of morphological features recomputed with skimage.measure.regionprops on the ROIs produced by ZooProcess. See http://scikit-image.org/docs/dev/api/skimage.measure.html#skimage.measure.regionprops for documentation. inventory.tsv Tree view of the taxonomy and number of images in each taxon, displayed as text. With columns : - lineage_level1: taxonomic lineage corresponding to the level 1 classification - taxon_level1: name of the taxon corresponding to the level 1 classification - n: number of objects in each taxon class map.png Map of the sampling locations, to give an idea of the diversity sampled in this dataset. imgs Directory containing images of each object, named according to the object id objid and sorted in subdirectories according to their taxon.
-
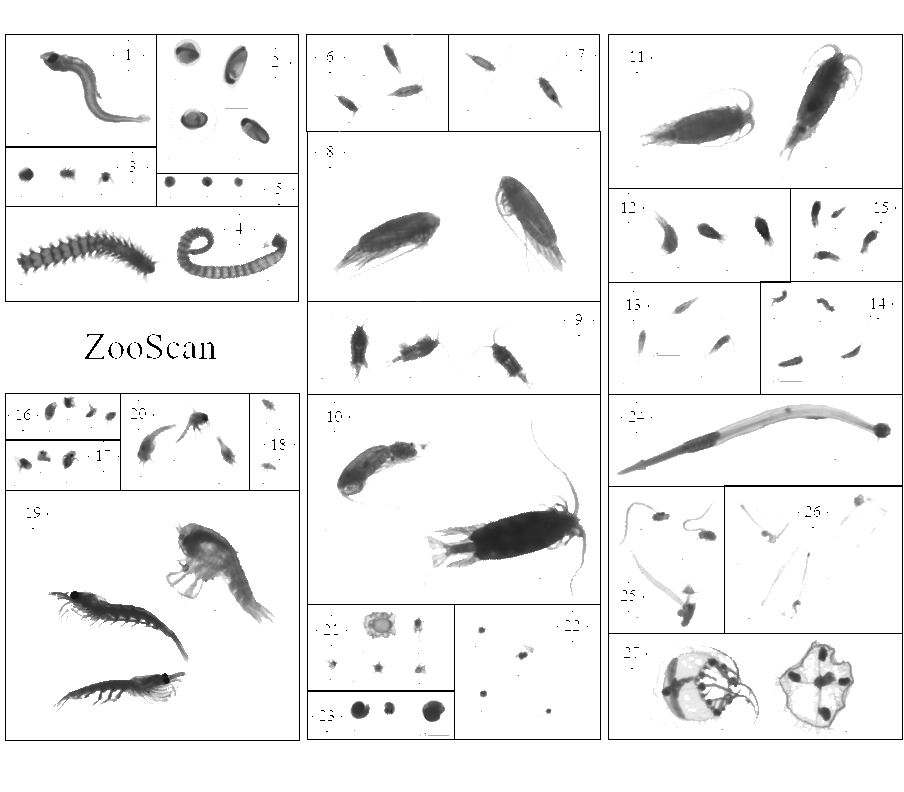
This dataset is composed of 1,153,507 zooplankton individuals, zooplankton parts, non-living particles and imaging artefacts, ranging from 300 µm to 3.39 mm Equivalent Spherical Diameter, individually imaged and measured with the ZooScan (Gorsky et al., 2010). The objects were sorted in 127 taxonomic and morphological groups. The imaged objects originate from samples collected on the Bay of Biscay continental shelf, in spring, from 2004 to 2016 during the PELGAS ecosystemic surveys (Doray et al., 2018). The samples were collected with a WP2 200 µm mesh size fitted with a Hydrobios (back-run stop) mechanical flowmeter, generally from 100 m depth to the surface, or 5 m above the sea floor (if bottom depth less than 100 m) in vertical hauls, at night. From 2004 to 2006, vertical WP2 net tows were performed in the anchovy core distribution area in the southern Bay of Biscay and North of it until the Loire estuary only. Since 2009, WP2 sampling has been carried out at all PELGAS stations, up to the southern coast of Brittany. The samples were preserved in 4% buffered formaldehyde seawater solution directly after collection, until 2019-2020 where they were imaged with the ZooScan, in the lab, on land. Each imaged object is geolocated, associated to a station, a cruise, a year and other metadata that enable the reconstruction of quantitative zooplankton communities for ecological studies (i.e. Grandrémy et al., 2023a). Each object is described by 46 morphological and grey level based features (8 bits encoding, 0 = black, 255 = white), including size, automatically extracted on each individual image by the Zooprocess. Each object was taxonomically identified using the web based application Ecotaxa with built-in, random forest and CNN based, semi-automatic sorting tools followed by expert validation or correction (Picheral et al., 2017). This dataset is intended to be used for ecological studies as well as machine learning applied to plankton studies. The archive contains: - One tab separated file (PELGAS ZooScan zooplankton dataset) containing all data and metadata associated to each imaged and identified object. Metadata and features are in columns (n =71) and objects are in rows (n = 1,153,507). - One comma separated file containing the name, type, definition and unit of each field (column) in the .tsv (dataset_descriptor_zooscan). - One comma separated file containing the taxonomic list of the dataset, with counts and nature of the content of the category, i.e. “T” for taxonomical category, and “M” for morphological category (taxonomy_descriptor_zooscan). - A individual_images directory containing images of each imaged object sorted in subdirectories named according to objects’ identifications object_taxon appended to an Ecotaxa internal taxon numerical id classif_id (i.e. taxon__123456789) across years and sampling stations. Within subdirectories, each object is named after its unique internal Ecotaxa identifier, objid. - A Map of the sampling station location over the 2004-2016 period
 Catalogue PIGMA
Catalogue PIGMA