plankton
Type of resources
Available actions
Topics
Keywords
Contact for the resource
Provided by
Years
Formats
Representation types
-
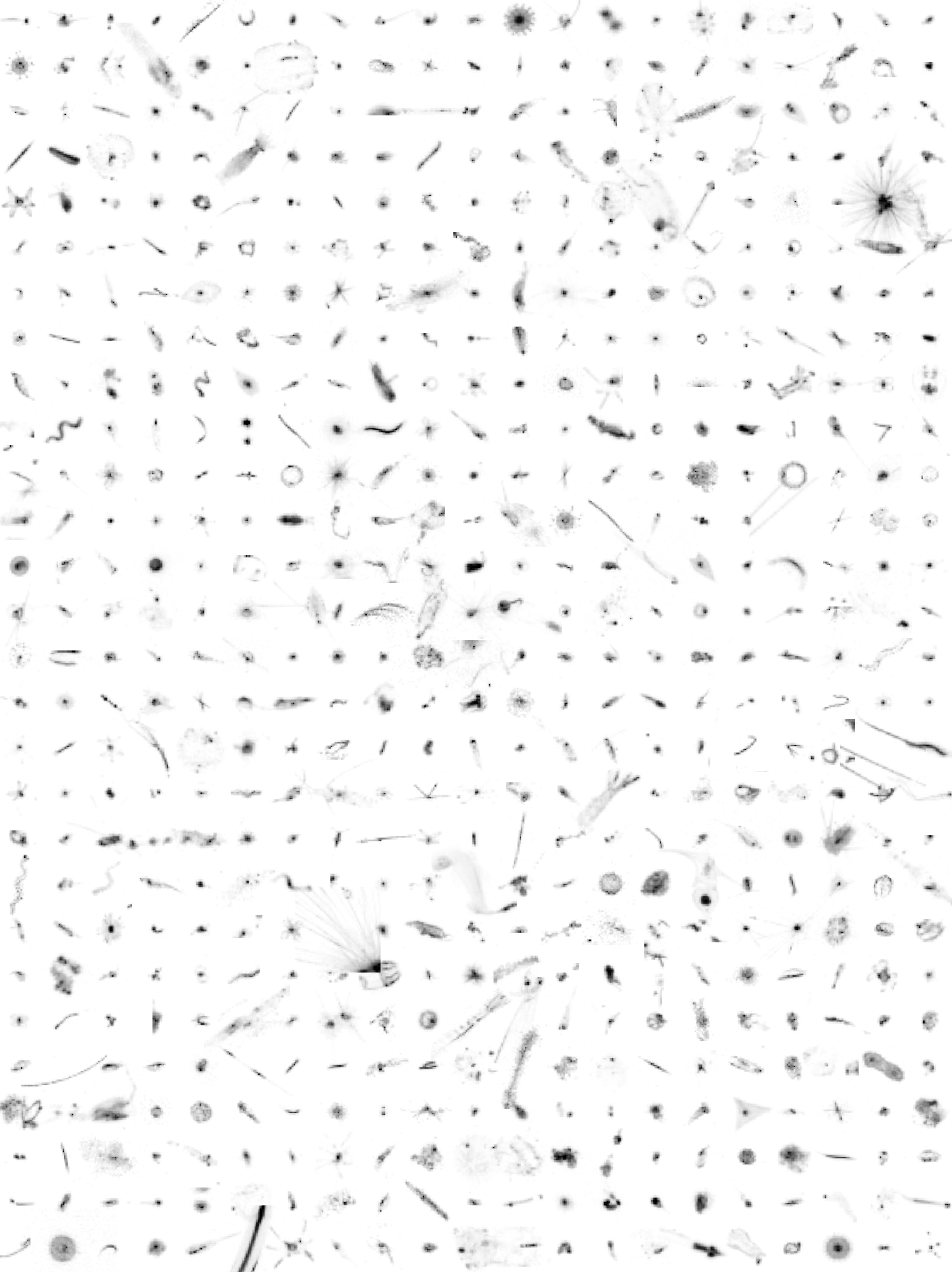
Here, we provide plankton image data that was sorted with the web applications EcoTaxa and MorphoCluster. The data set was used for image classification tasks as described in Schröder et. al (in preparation) and does not include any geospatial or temporal meta-data. Plankton was imaged using the Underwater Vision Profiler 5 (Picheral et al. 2010) in various regions of the world's oceans between 2012-10-24 and 2017-08-08. This data publication consists of an archive containing "training.csv" (list of 392k training images for classification, validated using EcoTaxa), "validation.csv" (list of 196k validation images for classification, validated using EcoTaxa), "unlabeld.csv" (list of 1M unlabeled images), "morphocluster.csv" (1.2M objects validated using MorphoCluster, a subset of "unlabeled.csv" and "validation.csv") and the image files themselves. The CSV files each contain the columns "object_id" (a unique ID), "image_fn" (the relative filename), and "label" (the assigned name). The training and validation sets were sorted into 65 classes using the web application EcoTaxa (http://ecotaxa.obs-vlfr.fr). This data shows a severe class imbalance; the 10% most populated classes contain more than 80% of the objects and the class sizes span four orders of magnitude. The validation set and a set of additional 1M unlabeled images were sorted during the first trial of MorphoCluster (https://github.com/morphocluster). The images in this data set were sampled during RV Meteor cruises M92, M93, M96, M97, M98, M105, M106, M107, M108, M116, M119, M121, M130, M131, M135, M136, M137 and M138, during RV Maria S Merian cruises MSM22, MSM23, MSM40 and MSM49, during the RV Polarstern cruise PS88b and during the FLUXES1 experiment with RV Sarmiento de Gamboa. The following people have contributed to the sorting of the image data on EcoTaxa: Rainer Kiko, Tristan Biard, Benjamin Blanc, Svenja Christiansen, Justine Courboules, Charlotte Eich, Jannik Faustmann, Christine Gawinski, Augustin Lafond, Aakash Panchal, Marc Picheral, Akanksha Singh and Helena Hauss In Schröder et al. (in preparation), the training set serves as a source for knowledge transfer in the training of the feature extractor. The classification using MorphoCluster was conducted by Rainer Kiko. Used labels are operational and not yet matched to respective EcoTaxa classes.
-
This dataset contains the pictures used for morphometric measurements, as well as the elemental compositon and production rates data, of planktonic Rhizaria. Specimens were collected in the bay of Villefranche-sur-Mer in May 2019 and during the P2107 cruise in the California Current in July-August 2021. Analyses of the data can be found at https://github.com/MnnLgt/Elemental_composition_Rhizaria.
-

Plankton was sampled with various nets, from bottom or 500m depth to the surface, in many oceans of the world. Samples were imaged with a ZooScan. The full images were processed with ZooProcess which generated regions of interest (ROIs) around each individual object and a set of associated features measured on the object (see Gorsky et al 2010 for more information). The same objects were re-processed to compute features with the scikit-image toolbox http://scikit-image.org. The 1,451,745 resulting objects were sorted by a limited number of operators, following a common taxonomic guide, into 98 taxa, using the web application EcoTaxa http://ecotaxa.obs-vlfr.fr. For the purpose of training machine learning classifiers, the images in each class were split into training, validation, and test sets, with proportions 70%, 15% and 15%. taxa.csv.gz Table of the classification of each object in the dataset, with columns : - objid: unique object identifier in EcoTaxa (integer number). - taxon_level1: taxonomic name corresponding to the level 1 classification - lineage_level1: taxonomic lineage corresponding to the level 1 classification - taxon_level2: name of the taxon corresponding to the level 2 classification - plankton: if the object is a plankton or not (boolean) - set: class of the image corresponding to the taxon (train : training, val : validation, or test) - img_path: local path of the image corresponding to the taxon (of level 1), named according to the object id features_native.csv.gz - objid: same as above - area: area - mean: mean grey - stddev: standard deviation of greys - mode: modal grey - min: minimum grey - max: maximum grey - perim.: perimeter - width,height dimensions - major,minor: length of major,minor axis of the best fitting ellipse - circ.: circularity: 4pi(area/perim.^2) - feret: maximal feret diameter - intden: integrated density: mean*area - median: median grey - skew,kurt: skewness,kurtosis of the histogram of greys - %area: proportion of the image corresponding to the object - area_exc: area excluding holes - fractal: fractal dimension of the perimeter - skelarea: area of the one-pixel wide skeleton of the image - slope: slope of the cumulated histogram of greys - histcum1,2,3: grey level at quantiles 0.25, 0.5, 0.75 of the histogram of greys - nb1,2,3: number of objects after thresholding at the grey levels above - symetrieh,symetriev: index of horizontal,vertical symmetry - symetriehc,symetrievc: same but after thresholding at level histcum1 - convperim,convarea: perimeter,area of the convex hull of the object - fcons: contrast - thickr: thickness ratio: maximum thickness/mean thickness - esd: Equivalent Spherical Diameter - elongation: elongation index: major/minor - range: range of greys: max-min - centroids : sqrt(pow(xm-x,2)+ pow(ym-y,2)) - sr: index of variation of greys: 100*(stddev/range) - perimareaexc : perim/(sqrt(area_exc)) - feretareaexc : feret/(sqrt(area_exc)) - perimferet: index of the relative complexity of the perimeter: perim/feret - perimmajor: index of the relative complexity of the perimeter: perim/major - circex: (4*PI*area_exc)/(pow(perim,2)) - cdexc: (1/(sqrt(area_exc))) * sqrt(pow(xm-x,2)+pow(ym-y,2) features_skimage.csv.gz Table of morphological features recomputed with skimage.measure.regionprops on the ROIs produced by ZooProcess. See http://scikit-image.org/docs/dev/api/skimage.measure.html#skimage.measure.regionprops for documentation. inventory.tsv Tree view of the taxonomy and number of images in each taxon, displayed as text. With columns : - lineage_level1: taxonomic lineage corresponding to the level 1 classification - taxon_level1: name of the taxon corresponding to the level 1 classification - n: number of objects in each taxon class map.png Map of the sampling locations, to give an idea of the diversity sampled in this dataset. imgs Directory containing images of each object, named according to the object id objid and sorted in subdirectories according to their taxon.
-
The European Ocean Biogeographic Information System - EurOBIS - is an online marine biogeographic database compiling data on all living marine creatures. The principle aims of EurOBIS are to centralize the largely scattered biogeographic data on marine species collected by European institutions and to make these data freely available and easily accessible. All data go through a number of quality control procedures before they are made available online, assuring a minimum level of quality necessary to put the data to good use. The available data are either collected within European marine waters or by European researchers and institutes outside Europe. The database focuses on taxonomy and distribution records in space and time; all data can be searched and visualised through a set of online mapping tools. All data are freely available online and easily accessible, without requiring a login or password.
-

EMODnet Biology provides three keys services and products to users. 1)The data download toolbox allows users to explore available datasets searching by source, geographical area, and/or time period. Datasets can be narrowed down using a taxonomic criteria, whether by species group (e.g. benthos, fish, algae, pigments) or by both scientific and common name. 2) The data catalogue is the easiest way to access nearly 1000 datasets available through EMODnet Biology. Datasets can be filtered by multiple parameters via the advanced search from taxon, to institute, to geographic region. Each of the resulting datasets then links to a detailed fact sheet containing a link to original data provider, recommended citation, policy and other relevant information. Data Products - EMODnet Biology combines different data from datasets with overlapping geographic scope and produces dynamic maps of selected species abundance. The first products are already available and they focus on species whose data records are most complete and span for a longer term.
-

The International Council for the Exploration of the Sea (ICES), is a global organization that develops science and advice to support the sustainable use of the oceans. ICES is a network of more than 5,000 scientists from over 690 marine institutes in 20 member countries and beyond. 1,500 scientists participate in our activities annually. ICES has a well-established Data Centre, which manages a number of large dataset collections related to the marine environment. The majority of data – covering the Northeast Atlantic, Baltic Sea, Greenland Sea, and Norwegian Sea – originate from national institutes that are part of the ICES network. The ICES Data Centre provides marine data services to ICES member countries, expert groups, world data centres, regional seas conventions (HELCOM and OSPAR), the European Environment Agency (EEA), Eurostat, and various other European projects and biodiversity portals. ICES aims to provide all data collections online and according to the ICES Data policy, which enables open access to all data that are do not fall under specific commercial or personal privacy concerns.
 Catalogue PIGMA
Catalogue PIGMA