2025
Type of resources
Available actions
Topics
Keywords
Contact for the resource
Provided by
Years
Formats
Representation types
Update frequencies
status
Service types
Scale
Resolution
-

Rocch, the french "mussel watch", provides chemical data for marine quality management. Once a year, trace metals, organic compounds (chlorinated, PAH, brominated flame retardants, perfluorinated compounds, organotins ...) are analysed in molluscs tissues to check chemical quality according to European Framework Directives and to Regional Seas Convention (OSPAR).
-
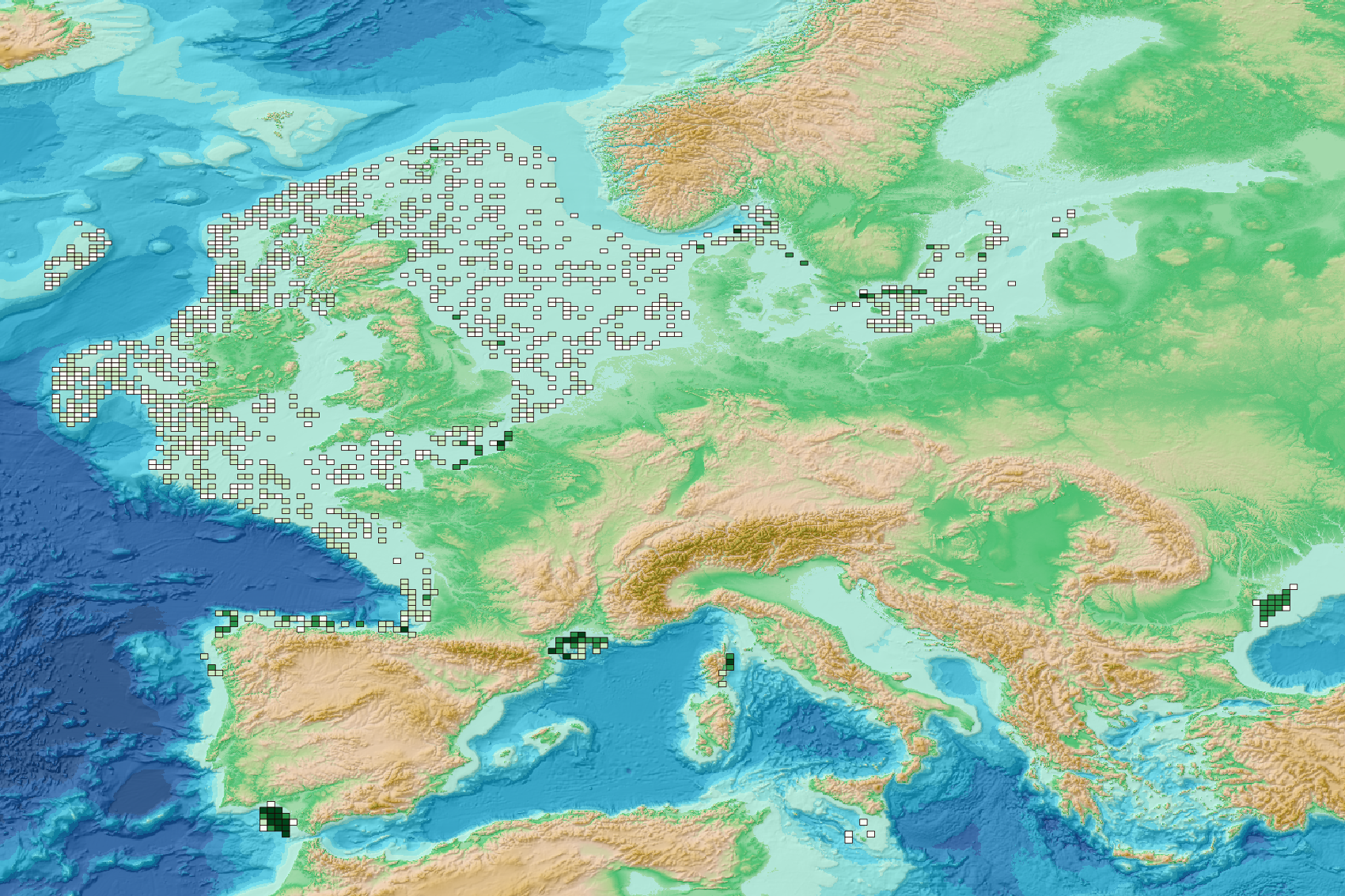
This visualization product displays the spatial distribution of the sampling effort over the six-years' period 2017-2022. EMODnet Chemistry included the collection of marine litter in its 3rd phase. Since the beginning of 2018, data of seafloor litter collected by international fish-trawl surveys have been gathered and processed in the EMODnet Chemistry Marine Litter Database (MLDB). The harmonization of all the data has been the most challenging task considering the heterogeneity of the data sources, sampling protocols (OSPAR and MEDITS protocols) and reference lists used on a European scale. Moreover, within the same protocol, different gear types are deployed during bottom trawl surveys. The spatial distribution was determined by calculating the number of times each cell was sampled during the period 2017-2022. The corresponding total distance (kms) sampled in each cell is also provided in the attribute table. Information on data processing and calculation are detailed in the attached methodology document. Warning: the absence of data on the map does not necessarily mean that they do not exist, but that no information has been entered in the Marine Litter Database for this area. This work is based on the work presented in the following scientific article: O. Gerigny, M. Brun, M.C. Fabri, C. Tomasino, M. Le Moigne, A. Jadaud, F. Galgani, Seafloor litter from the continental shelf and canyons in French Mediterranean Water: Distribution, typologies and trends, Marine Pollution Bulletin, Volume 146, 2019, Pages 653-666, ISSN 0025-326X, https://doi.org/10.1016/j.marpolbul.2019.07.030.
-

The network was initiated by IFREMER from 1993 to 2009 (under the acronym REMORA) to study the rearing performance of the Pacific oyster Crassostrea gigas at a national scale. To do so, the network monitored annually the mortality and growth of standardized batches of 18-month-old oysters. Starting in 1995, the monitoring of the rearing performance of 6-month-old oyster spat was integrated into this network. These sentinel batches were distributed simultaneously each year on 43 sites and were monitored quarterly. These sites were distributed over the main French oyster farming areas and allowed a national coverage of the multiannual evolution of oyster farming performances. Most of the sites were located on the foreshore at comparable levels of immersion. Field studies were carried out by the "Laboratoires Environnement Ressources" (LER) for the sites included in their geographical area of investigation. Following the increase in spat mortality in 2008, the network evolved in 2009 (under the acronym RESCO). From this date, the network selected 13 sites among the 43 sites previously monitored in order to increase the frequency of visits (twice a month) and the number of sentinel batches. More precisely, sentinel batches of oysters corresponding to different origins (wild or hatchery, diploid or triploid) and to two rearing age classes (spat or 18-month-old adults) were selected. The monitoring of environmental variables (temperature, salinity) associated with the 13 sites was also implemented. The actions of the network have thus contributed to disentangle the biotic and abiotic parameters involved in mortality phenomena, taking into account the different compartments (environment / host / infectious agents) likely to interact with the evolution of oyster rearing performance. Finally, since 2015, the network has merged the RESCO and VELYGER networks to adopt the acronym ECOSCOPA. The general objective of this current network is to analyze the causes of spatio-temporal variability of the main life traits (Larval stage - Recruitment - Reproduction - Growth - Survival - Cytogenetic abnormalities) of the cupped oyster in France and to follow their evolution on the long term in the context of climate change. To do this, the network proposes a regular spatio-temporal monitoring of the major proxies of the life cycle of the oyster, organized in three major thematic groups: (1) proxies related to growth, physiological tolerance and survival of experimental sentinel populations over 3 age classes: (2) proxies related to reproduction, larval phase and recruitment of the species throughout its natural range in France, and: (3) proxies related to environmental parameters essential to the species (weather conditions, temperature, salinity, pH, turbidity, chlorophyll a and phytoplankton) at daily or sub-hourly frequencies. Working in a geographical network associating several laboratories, ECOSCOPA provide these monitoring within 8 sites selected among the previous ones to ensure the continuity of the data acquisition. Today, these 8 sites are considered as ecosystems of common interest, contrasted, namely : - The Thau lagoon - The Arcachon basin - The Marennes Oléron basin - The Bourgneuf Bay - The bay of Vilaine - The bay of Brest - The bay of Mont Saint Michel - The bay of Veys The ECOSCOPA network is therefore one of the relevant monitoring tools on a national scale, allowing to objectively measure through different proxies the general state of health of cultivated and wild oyster populations, and this for the different sensitive phases of their life cycle. This network aims at allowing a better evaluation, on the long term, of the biological risks incurred by the sector but also by the ecosystems, in particular under the increasing constraint of climatic and anthropic changes. Figure : Sites monitored by the ECOSCOPA network
-

EMODnet Chemistry aims to provide access to marine chemistry datasets and derived data products concerning eutrophication, acidity and contaminants. The importance of the selected substances and other parameters relates to the Marine Strategy Framework Directive (MSFD). This aggregated dataset contains all unrestricted EMODnet Chemistry data on eutrophication and acidity, and covers the Greater North Sea and Celtic Seas. Data were aggregated and quality controlled by 'Aarhus University, Department of Bioscience, Marine Ecology Roskilde' in Denmark. ITS-90 water temperature and water body salinity variables have also been included ('as are') to complete the eutrophication and acidity data. If you use these variables for calculations, please refer to SeaDataNet for the quality flags: https://www.seadatanet.org/Products/Aggregated-datasets . Regional datasets concerning eutrophication and acidity are automatically harvested, and the resulting collections are aggregated and quality controlled using ODV Software and following a common methodology for all sea regions ( https://doi.org/10.13120/8xm0-5m67 ). Parameter names are based on P35 vocabulary, which relates to EMODnet Chemistry aggregated parameter names and is available at: https://vocab.nerc.ac.uk/search_nvs/P35/ . When not present in original data, water body nitrate plus nitrite was calculated by summing all nitrate and nitrite parameters. The same procedure was applied for water body dissolved inorganic nitrogen (DIN), which was calculated by summing all nitrate, nitrite, and ammonium parameters. Concentrations per unit mass were converted to a unit volume using a constant density of 1.025 kg/L. The aggregated dataset can also be downloaded as an ODV collection and spreadsheet, which is composed of a metadata header followed by tab separated values. This spreadsheet can be imported to ODV Software for visualisation (more information can be found at: https://www.seadatanet.org/Software/ODV ).
-
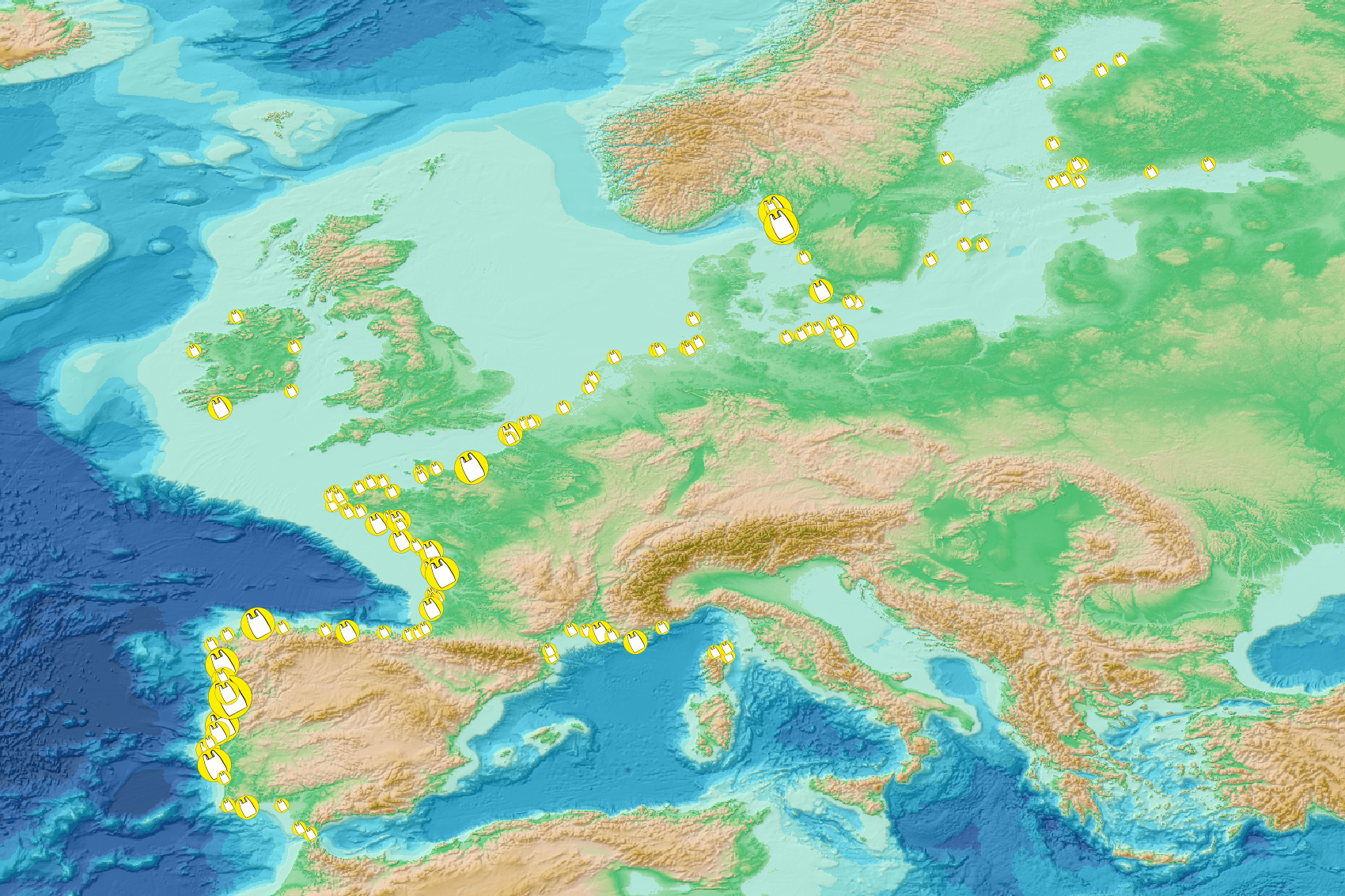
This visualization product displays the fishing & aquaculture related plastic items abundance of marine macro-litter (> 2.5cm) per beach per year from Marine Strategy Framework Directive (MSFD) monitoring surveys. EMODnet Chemistry included the collection of marine litter in its 3rd phase. Since the beginning of 2018, data of beach litter have been gathered and processed in the EMODnet Chemistry Marine Litter Database (MLDB). The harmonization of all the data has been the most challenging task considering the heterogeneity of the data sources, sampling protocols and reference lists used on a European scale. Preliminary processings were necessary to harmonize all the data: - Exclusion of OSPAR 1000 protocol: in order to follow the approach of OSPAR that it is not including these data anymore in the monitoring; - Selection of MSFD surveys only (exclusion of other monitoring, cleaning and research operations); - Exclusion of beaches without coordinates; - Selection of plastic bags related items only. The list of selected items is attached to this metadata. This list was created using EU Marine Beach Litter Baselines, the European Threshold Value for Macro Litter on Coastlines and the Joint list of litter categories for marine macro-litter monitoring from JRC (these three documents are attached to this metadata); - Normalization of survey lengths to 100m & 1 survey / year: in some case, the survey length was not exactly 100m, so in order to be able to compare the abundance of litter from different beaches a normalization is applied using this formula: Number of plastic bags related items of the survey (normalized by 100 m) = Number of plastic bags related items of the survey x (100 / survey length) Then, this normalized number of plastic bags related items is summed to obtain the total normalized number of plastic bags related items for each survey. Finally, the median abundance of plastic bags related items for each beach and year is calculated from these normalized abundances of plastic bags related items per survey. Sometimes the survey length was null or equal to 0. Assuming that the MSFD protocol has been applied, the length has been set at 100m in these cases. Percentiles 50, 75, 95 & 99 have been calculated taking into account plastic bags related items from MSFD data for all years. More information is available in the attached documents. Warning: the absence of data on the map does not necessarily mean that they do not exist, but that no information has been entered in the Marine Litter Database for this area.
-

Particularly suited to the purpose of measuring the sensitivity of benthic communities to trawling, a trawl disturbance indicator (de Juan and Demestre, 2012, de Juan et al. 2009) was proposed based on benthic species life history traits to evaluate the sensibility of mega- and epifaunal community to fishing pressure known to have a physical impact on the seafloor (such as dredging and bottom trawling). The selected biological traits were chosen as they determine vulnerability to trawling: mobility, fragility, position on substrata, average size and feeding mode that can easily be related to the fragility, recoverability and vulnerability ecological concepts. Life history traits of species have been defined from the BIOTIC database (MARLIN, 2014) and from information given by Le Pape et al. (2007), Brindamour et al. (2009) and Garcia (2010). For missing life history traits, additional information from literature has been considered. The five categories retained are life history functional traits that were selected based on the knowledge of the response of benthic taxa to trawling disturbance (de Juan and Demestre, 2012). They reflect respectively the possibility to avoid direct gear impact, to benefit from trawling for feeding, to escape gear, to get caught by the net and to resist trawling/dredging action, each of these characteristics being either advantageous or sensitive to trawling. Then, to allow quantitative analysis, a score was assigned to each category: from low vulnerability (0) to high vulnerability (3). The five categories scores were then summed for each taxon (the highly vulnerable taxon could reach the maximum score is 15) and this value may be considered as a species index of sensitivity to trawling disturbance. The scores of 812 taxa commonly found in bottom trawl by-catch in the southern North Sea, English Channel and north-western Mediterranean were described.
-
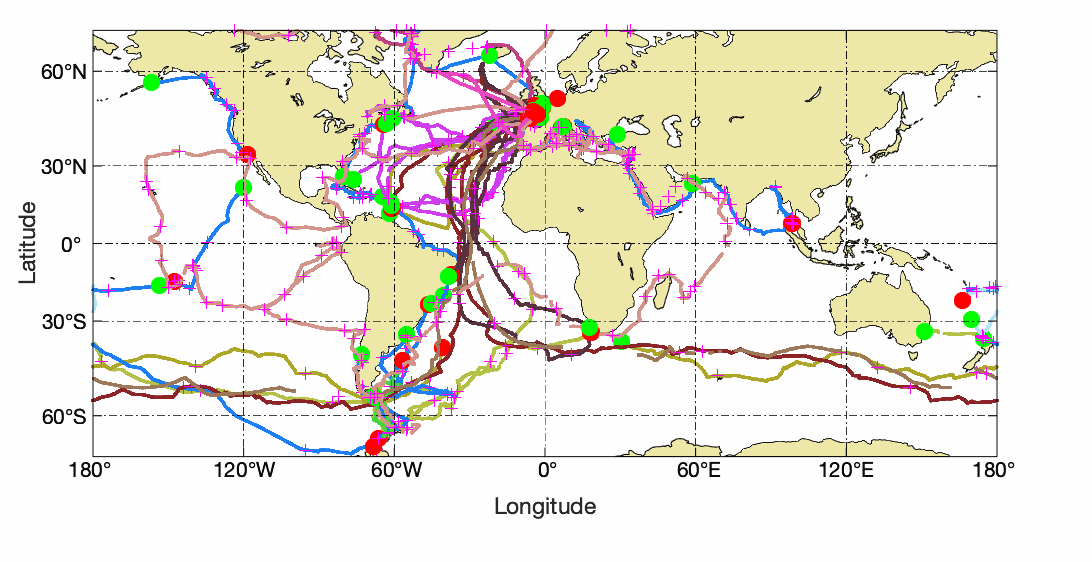
Observations of Sea surface temperature and salinity are now obtained from voluntary sailing ships using medium or small size sensors. They complement the networks installed on research vessels or commercial ships. The delayed mode dataset proposed here is upgraded annually as a contribution to GOSUD (http://www.gosud.org )
-

Numerous reef-forming species have declined dramatically in the last century, many of which have been insufficiently documented due to anecdotal or hard-to-access information. One of them, the honeycomb worm Sabellaria alveolata (L.) is a tube-building polychaete that can form large reefs, providing important ecosystem services such as coastal protection and habitat provision. It ranges from Scotland to Morocco, yet little is known about its distribution outside of the United Kingdom, where it is protected and where there is a strong heritage of natural history and sustained observations. As a result, online marine biodiversity information systems currently contain haphazardly distributed records of S. alveolata. One of the objectives of the REEHAB project (http://www.honeycombworms.org) was to combine historical records with contemporary data to document changes in the distribution and abundance of S. alveolata. Here we publish the result of the curation of 446 sources, gathered from literature, targeted surveys, local conservation reports, museum specimens, personal communications by authors and by their research teams, national biodiversity information systems (i.e. the UK National Biodiversity Network (NBN), https://nbn.org.uk/) and validated citizen science observations (i.e. https://www.inaturalist.org/). 80%[ar1] of these records were not previously referenced in any online information system. Additionally, historic field notebooks from Edouard Fischer-Piette and Gustave Gilson were scanned for S. alveolata information and manually entered. The original taxonomic identification of the 23296 S. alveolata records has been kept. Some identification errors may however be present, particularly in the English Channel and the North Sea where incorrectly identified observations of intertidal Sabellaria spinulosa were recorded. A further 229 observations are recorded as ‘Sabellaria spp.’ as the available information does not allow a species-level identification. Many sources reported abundances based on the semi-quantitative SACFOR scale while others simply noted its presence, and others still verified both its absence and presence. The result is a curated and comprehensive dataset spanning over two centuries on the past and present global distribution and abundance of S. alveolata. Sabellaria alveolata records projected onto a 50km grid. When SACFOR scale abundance scores were given to occurrence records, the highest abundance value per grid cell was retained.
-
The raster corresponds to the predicted Mediterranean bioregions of megabenthic communities.
-
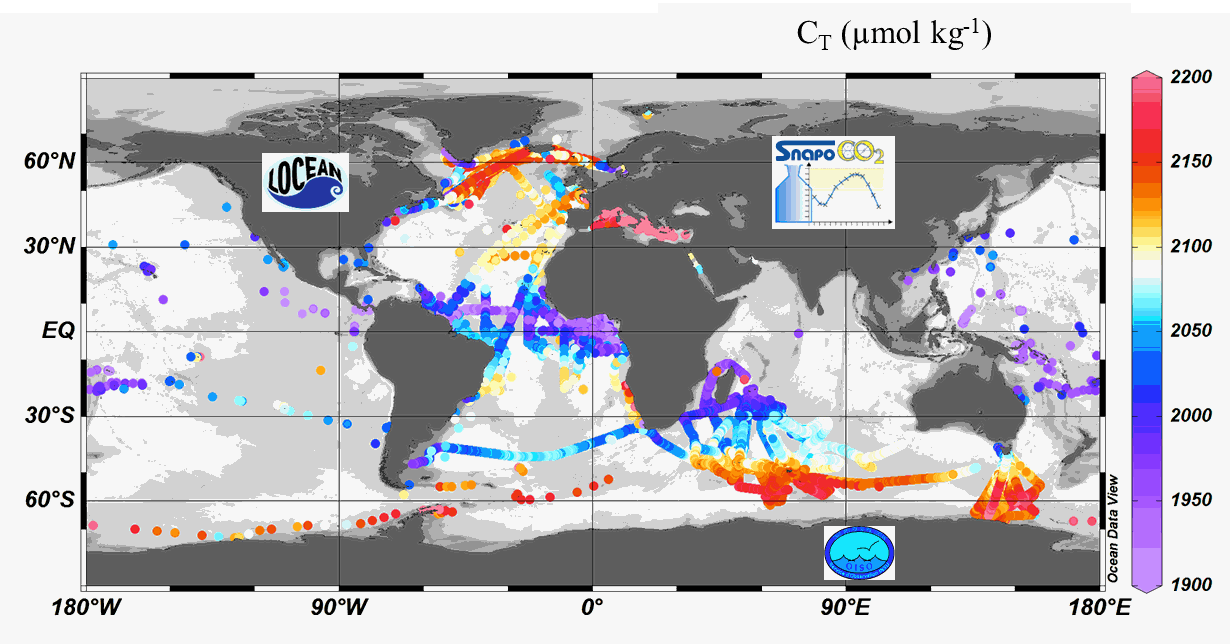
Since 2004, the Service facility SNAPO-CO2 (Service National d’Analyse des Paramètres Océaniques du CO2) housed by the LOCEAN laboratory (Paris, France) has been in charge for the analysis of Total Alkalinity (AT) and Total dissolved inorganic carbon (CT) of seawater samples on a series of cruises or ships of opportunity conducted in different regions in the frame of French and International projects. Following the first synthesis (Metzl et al, 2024, https://doi.org/10.17882/95414), 24700 new data have been quality controlled and the second version includes more than 67000 observations over 1993-2023. Sampling was performed either from CTD-Rosette casts (Niskin bottles) or collected from the ship’s seawater supply (intake at about 5m depth). After completion of each cruise, discrete samples were returned back at LOCEAN laboratory and stored in a dark room at 4 °C before analysis generally within 2-3 months after sampling (sometimes within a week). AT and CT were analyzed simultaneously by potentiometric titration using a closed cell (Edmond, 1970). Certified Reference Materials (CRMs) provided by Pr. A. Dickson (Scripps Institution of Oceanography, San Diego, USA) were used to calibrate the measurements. The same instrumentation was used for underway measurements during OISO cruises (https://campagnes.flotteoceanographique.fr/series/228/) and MINERVE cruises (https://doi.org/10.18142/128) and new AT-CT data for 2002-2021 in the Indian Ocean and Southern Ocean added in this synthesis. The second dataset is organized in one file with the format: Cruise name, Ship name, day, month, year, hour, minute, second, latitude, longitude, depth, AT (µmol/kg), Flag-AT, CT (µmol/kg), Flag-CT, Temperature (°C), Flag-Temp, Salinity (PSU), Flag-Salinity, nsample/cruise, sampling method, Version number, nsample on file.
 Catalogue PIGMA
Catalogue PIGMA