CSV
Type of resources
Available actions
Topics
Keywords
Contact for the resource
Provided by
Years
Formats
Representation types
Update frequencies
status
Scale
Resolution
-

This folder contains two examples of PAGURE datasets, corresponding to three surveys: -CGFS conducted in 2018 in the English Channel (Northeast Atlantic) -EPIBENGOL conducted in 2019 in the Gulf of Lion (Western Mediterranean) -EVHOE conducted in 2020 in the Bay of Biscay and Celtic Shelf (Northeast Atlantic) Files include metadata for the sampling stations, annotation files. A readme tex file contains the links to the voyage metadata This folder is aimed at providing an example of documented underwater imagery dataset. These data are part of the data exchange conducted in the QuatreA collaboration between the French Research Institute for the Exploitation of the Sea (Ifremer), the Commonwealth Scientific and Industrial Research Organisation (CSIRO), and the University of Tasmania (UTAS).
-

The network was initiated by IFREMER from 1993 to 2009 (under the acronym REMORA) to study the rearing performance of the Pacific oyster Crassostrea gigas at a national scale. To do so, the network monitored annually the mortality and growth of standardized batches of 18-month-old oysters. Starting in 1995, the monitoring of the rearing performance of 6-month-old oyster spat was integrated into this network. These sentinel batches were distributed simultaneously each year on 43 sites and were monitored quarterly. These sites were distributed over the main French oyster farming areas and allowed a national coverage of the multiannual evolution of oyster farming performances. Most of the sites were located on the foreshore at comparable levels of immersion. Field studies were carried out by the "Laboratoires Environnement Ressources" (LER) for the sites included in their geographical area of investigation. Following the increase in spat mortality in 2008, the network evolved in 2009 (under the acronym RESCO). From this date, the network selected 13 sites among the 43 sites previously monitored in order to increase the frequency of visits (twice a month) and the number of sentinel batches. More precisely, sentinel batches of oysters corresponding to different origins (wild or hatchery, diploid or triploid) and to two rearing age classes (spat or 18-month-old adults) were selected. The monitoring of environmental variables (temperature, salinity) associated with the 13 sites was also implemented. The actions of the network have thus contributed to disentangle the biotic and abiotic parameters involved in mortality phenomena, taking into account the different compartments (environment / host / infectious agents) likely to interact with the evolution of oyster rearing performance. Finally, since 2015, the network has merged the RESCO and VELYGER networks to adopt the acronym ECOSCOPA. The general objective of this current network is to analyze the causes of spatio-temporal variability of the main life traits (Larval stage - Recruitment - Reproduction - Growth - Survival - Cytogenetic abnormalities) of the cupped oyster in France and to follow their evolution on the long term in the context of climate change. To do this, the network proposes a regular spatio-temporal monitoring of the major proxies of the life cycle of the oyster, organized in three major thematic groups: (1) proxies related to growth, physiological tolerance and survival of experimental sentinel populations over 3 age classes: (2) proxies related to reproduction, larval phase and recruitment of the species throughout its natural range in France, and: (3) proxies related to environmental parameters essential to the species (weather conditions, temperature, salinity, pH, turbidity, chlorophyll a and phytoplankton) at daily or sub-hourly frequencies. Working in a geographical network associating several laboratories, ECOSCOPA provide these monitoring within 8 sites selected among the previous ones to ensure the continuity of the data acquisition. Today, these 8 sites are considered as ecosystems of common interest, contrasted, namely : - The Thau lagoon - The Arcachon basin - The Marennes Oléron basin - The Bourgneuf Bay - The bay of Vilaine - The bay of Brest - The bay of Mont Saint Michel - The bay of Veys The ECOSCOPA network is therefore one of the relevant monitoring tools on a national scale, allowing to objectively measure through different proxies the general state of health of cultivated and wild oyster populations, and this for the different sensitive phases of their life cycle. This network aims at allowing a better evaluation, on the long term, of the biological risks incurred by the sector but also by the ecosystems, in particular under the increasing constraint of climatic and anthropic changes. Figure : Sites monitored by the ECOSCOPA network
-

-
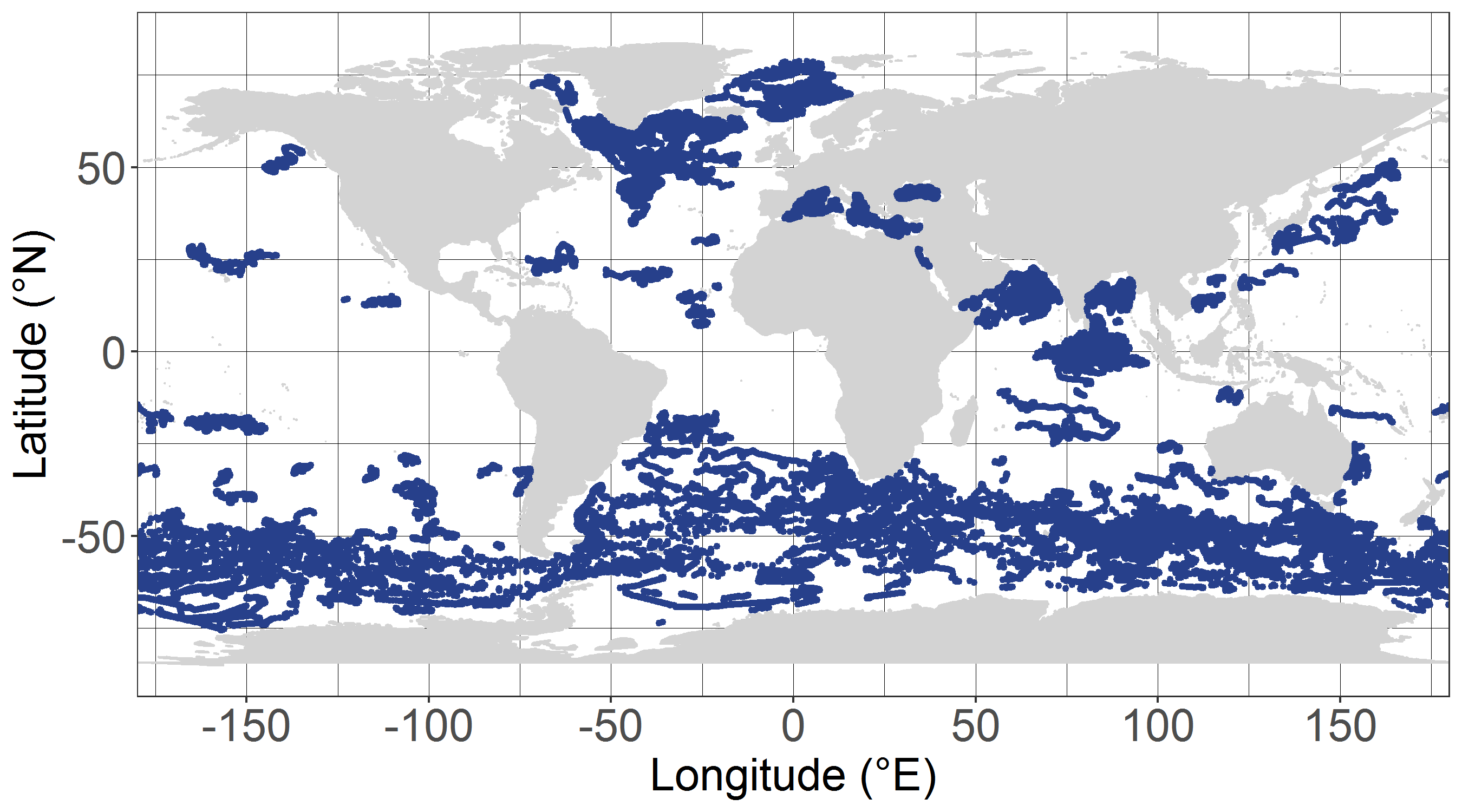
The dataset dcm_dtb.txt contains bio-optical measurements and environmental parameters associated with Deep Chlorophyll Maxima (DCM) acquired by BGC-Argo profiling floats. For each BGC-Argo profile the data files includes the World Meteorological Organization (WMO) and profile numbers, the Data Assembly Center (DAC), the geographical position (LON and LAT), the date of the profile in Julian Day (JULD) and in YYYY-MM-DD format; the region of the profile (REGION, acronyms detailed in the region.txt file), the DCM zonal attribution (ZONE, acronyms detailed in the zone.txt file), the vertical resolution of measurements of the concentration of the chlorophyll a [Chla] and of the backscattering coefficient (bbp) within the 250 first meters, the Mixed Layer Depth (MLD, m), the qualification of the vertical profile (DCM_TYPE) as Deep Biomass Maximum (3), Deep photoAcclimation Maximum (2), or presenting no DCM (1); the depth of the DCM (DCM_DEPTH); the chlorophyll a concentration (CHLA_DCM, mg chla m-3 ) the backscattering coefficient (BBP_DCM, m-1), and the Brunt-Vaisala frequency (N2_DCM) at the DCM depth; the nitracline depth (NCLINE_DEPTH, m) and steepness (NCLINE_STEEP, µmol NO3 m-3 m-1), the mean nitrate concentration within the Mixed Layer (NO3_MEAN_MLD, µmol NO3 m-3), the mean daily Photosynthetically Available Radiation in the Mixed Layer (MEAN_IPAR_MLD, E m -1 d -1), the daily Photosynthetically Available Radiation at the nitracline depth (IPAR_NCLINE, E m-2 d-1); and the [Chla] measured by satellite (CHLA_SAT, mg chla m-3). The dataset shape_NASTG_ASEW.txt contains the seasonal median, the first and third quartiles of the [Chla] and of the bbp profiles for the North Atlantic Subtropical Gyre and Atlantic SubEquatorial Waters regions. The dataset climato_NASTG_ASEW.txt contains the monthly mean and standard deviations of the DCM depth (DCM_depth), the isolume depth of daily Photosynthetically Available Radiation of 20 E m-2 d-1 (iPAR_20), the nitracline depth, and the Mixed Layer Depth (MLD) for the profiles within the North Atlantic Subtropical Gyre and Atlantic SubEquatorial Waters regions. The qualification and processing of the BGC-Argo profiles, as well as the DCM detection (DCM_TYPE) and the estimation of the environmental parameters, were applied as described from Cornec, M., Claustre, H., Mignot, A., Guidi, L., Lacour, L., Poteau, A., D’Ortenzio, F.,Gentili, B., Schmechtig, C., (to be updated.) Deep Chlorophyll Maxima in the global ocean: occurrences, drivers and characteristics. Global Biogeochemical Cycles, to be updated The [Chla] satellite variable was obtained by the match of each BGC-Argo profile with a L3S [Chla] product from the Ocean Colour-Climate Change Initiative v4.0 database merging observations from MERIS, MODIS, VIIRS and SeaWiFs, at a monthly and 4x4-km-pixel resolution, up to December 31, 2019 (ftp://oc-cci-data:ELaiWai8ae@oceancolour.org/occci-v4.2/).
-
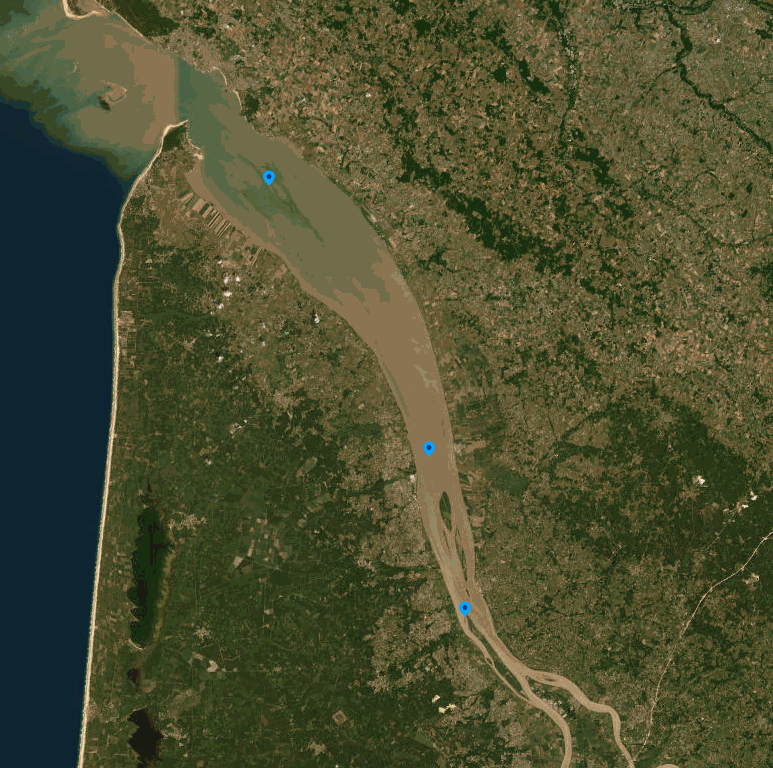
The SOMLIT-SOGIR time-series data characterize the hydrology of the Gironde Estuary ecosystem, located in the South-western France and flowing into the Bay of Biscay. Monthly-like measurements have been undertaken since 1997 by the OASU and EPOC teams (Univ. Bordeaux/CNRS). The SOMLIT-SOGIR time series is a part of the French monitoring network SOMLIT (https://www.somlit.fr/), labelled by the CNRS as a national Earth Science Observatory (Service National d’Observation : SNO). It aims to detect the long-term evolution of monitored ecosystems including both natural and anthropogenic forcings. Implemented at three sites (PK 30: 45.06833°N, 0.63833°W; PK 52: 45.24667°N, 0.725°W; PK 86: 45.5167°N, 0.95°W), the SOMLIT-SOGIR time series is among the oldest long-term coastal observation time series of the French Research Infrastructure dedicated to coastal ocean observations (RI ILICO, https://www.ir-ilico.fr). SOMLIT-SOGIR samples are collected at 1m below the water surface and 1m above the floor, at high and low tide, during slack water. Samples collected are analysed for 15 core parameters: water temperature and salinity, dissolved oxygen, pH, ammonia, nitrate, nitrite, phosphate, silicic acid, suspended particulate matter, particulate organic carbone, particulate nitrogen, chlorophyll a, delta15N and delta13C. CTD-PAR-profile is also performed at site PK86 during high tide. The SOMLIT network quality management system is in line with the ISO/IEC 17025:2017 standard: “General requirements for the competence of testing and calibration laboratories”. Further information on standard operating procedures for sample collection and data acquisition are available at: https://www.somlit.fr/parametres-et-protocoles. For more information on the quality flagging scheme: https://www.somlit.fr/codes-qualite/.
-
The COAST-HF/Arcachon-Ferret time series characterizes the hydrology of the interface between the Arcachon lagoon, located in the South-Western France, and the Atlantic Ocean. A buoy belonging to Phares et Balises is instrumented with a multi-parametric probe that records sub-surface temperature, conductivity, depth, turbidity and fluorescence every 10 minutes since February 2018. It is opérated by the OASU and EPOC teams (Univ. Bordeaux/CNRS). COAST-HF (Coastal OceAn observing SysTem - High Frequency; www.coast-hf.fr) is a national observation network accredited by the CNRS as a national Earth Science Observatory (Service National d’Observation: SNO). It aims to federate and coordinate a set of 14 fixed platforms instrumented with high-frequency in situ measurements for key parameters of coastal waters. The COAST-HF/Arcachon-Ferret buoy is one of them. COAST-HF is part of the French Research Infrastructure dedicated to coastal ocean observations (RI ILICO, https://www.ir-ilico.fr). Data are transmitted to the Coriolis Côtier database (https://data.coriolis-cotier.org/). Data are raw data.
-
The French Atlantic coast hosts numerous macrotidal and turbid estuaries that flow into the Bay of Biscay that are natural corridors for migratory fishes. The two best known are those of the Gironde and the Loire. However, there are also a dozen estuaries set geographically among them, of a smaller scale. The physico-chemical quality of estuarine waters is a necessary support element for biological life and determines the distribution of species, on which many ecosystem services (e.g. professional or recreational fishing) depend. With rising temperatures and water levels, declining precipitation and population growth projected for the New Aquitaine region by 2030, the question of how the quality and ecological status of estuarine waters will evolve becomes increasingly critical. The MAGEST (Mesures Automatisées pour l’observation et la Gestion des ESTuaires nord aquitains) high-frequency monitoring of key physico-chemical parameters was first developed in the Gironde estuary in 2004 ; the Seudre and Charente estuaries were instrumented late 2020. First based on real-time automated systems, MAGEST is now equipped by autonomous multiparameter sensors. Depending of the stations, an optode is also deployed to secure dissolved oxygen measurement. By the end of 2020, MAGEST had 12 instrumented sites. Portets is a measuring station located in the upper Gironde estuary (Garonne subestuary, about 20 km upstream of the Bordeaux metropolis.
-

The SOMLIT-Antioche observation station, located at 5 nautical miles from Chef de Baie harbor (La Rochelle) is part of the French monitoring network SOMLIT (https://www.somlit.fr/), accredited by the INSU-CNRS as a national Earth Science Observatory (Service National d’Observation : SNO), which comprises 12 observation stations distributed throughout France in coastal locations. It aims to detect long-term changes of these ecosystems under both natural and anthropogenic forcings. SOMLIT is part of the national research infrastructure for coastal ocean observation ILICO (https://www.ir-ilico.fr/?PagePrincipale&lang=en). The SOMLIT-Antioche station (46.0842 °N, 1.30833 °W) is located in the north-eastern part of the Bay of Biscay, halfway between the islands of Ré and Oléron, at the centre of what is commonly known as the Pertuis Charentais area, which correspond to a semi-enclosed shallow basin and includes four islands (Ré, Oléron, Aix and Madame) and three Pertuis (i.e., detroit) (Breton, Antioche and Maumusson). This 40m-deep site, with muddy to sandy marine bottoms, is submitted to a macro-tidal regime and is largely open to the prevailing westerly swells. It remains under a dominant oceanic/neritic influence, even though its winter/spring hydrological context is influenced by the diluted plumes of the Charente, Gironde and Loire rivers, but not by those of too small estuaries (Lay, Seudre and Sèvre Niortaise). SOMLIT-Antioche hydrological monitoring has been carried out by the LIENSs/OASU laboratory on a fortnightly basis since June 2011. Surface water samples are collected at high-tide during intermediate tides (70 ± 10 in SHOM units) on board the research vessel ‘L’Estran’ owned by La Rochelle University. Samples are analyzed for more than 16 core parameters: temperature, salinity, dissolved oxygen, pH, ammonia, nitrates, nitrites, phosphates, silicates, suspended matter, particulate organic carbone, particulate organic nitrogen, chlorophyll, delta15N, delta13C; pico- and nano- plankton. Measurements are carried out in accordance with the ISO/IEC 17025:2017 standard. Simultaneous monitoring of the micro-phytoplankton community (since 2013, SNO PHYTOBS: https://www.phytobs.fr/en) and monitoring of prokaryotic communities (Bacteria and Archaea) are also carried out on a monthly basis. Since 2019, seasonal observations of benthic invertebrate communities (SNO BenthObs : https://www.benthobs.fr/) have also been carried out. This monitoring is complementary to that carried out at hydrological stations in the pre-existing REPHY and DCE networks, some of which are located near marine farming areas (oyster and mussel farms).
-

Long-term time series of coliform bacteria concentration (fecal coliform or Escherichia coli) in shellfish in four submarine areas (North Sea/Channel, Britany, Atlantic, Mediterranean).
-
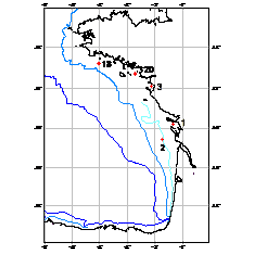
210Pb, 226Ra and 137Cs were measured by non-destructive gamma spectrometry on marine sediment cores, collected during RIKEAU 2002 cruise on board r/v Thalia, on the shelf of the Bay of Biscay
 Catalogue PIGMA
Catalogue PIGMA