IMAGE
Type of resources
Available actions
Topics
Keywords
Contact for the resource
Provided by
Years
Formats
-

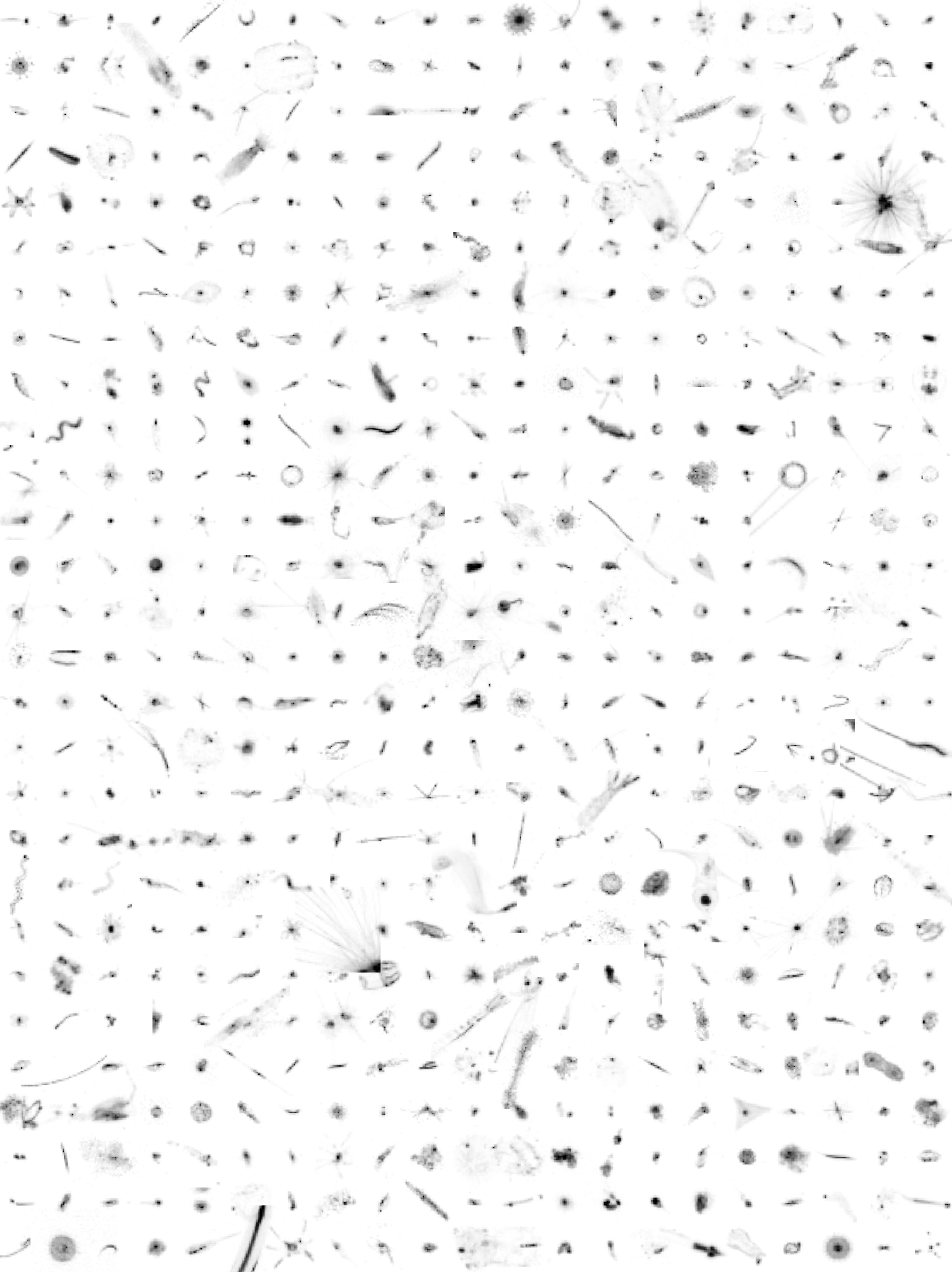
Plankton was sampled with various nets, from bottom or 500m depth to the surface, in many oceans of the world. Samples were imaged with a ZooScan. The full images were processed with ZooProcess which generated regions of interest (ROIs) around each individual object and a set of associated features measured on the object (see Gorsky et al 2010 for more information). The same objects were re-processed to compute features with the scikit-image toolbox http://scikit-image.org. The 1,451,745 resulting objects were sorted by a limited number of operators, following a common taxonomic guide, into 98 taxa, using the web application EcoTaxa http://ecotaxa.obs-vlfr.fr. For the purpose of training machine learning classifiers, the images in each class were split into training, validation, and test sets, with proportions 70%, 15% and 15%. taxa.csv.gz Table of the classification of each object in the dataset, with columns : - objid: unique object identifier in EcoTaxa (integer number). - taxon_level1: taxonomic name corresponding to the level 1 classification - lineage_level1: taxonomic lineage corresponding to the level 1 classification - taxon_level2: name of the taxon corresponding to the level 2 classification - plankton: if the object is a plankton or not (boolean) - set: class of the image corresponding to the taxon (train : training, val : validation, or test) - img_path: local path of the image corresponding to the taxon (of level 1), named according to the object id features_native.csv.gz - objid: same as above - area: area - mean: mean grey - stddev: standard deviation of greys - mode: modal grey - min: minimum grey - max: maximum grey - perim.: perimeter - width,height dimensions - major,minor: length of major,minor axis of the best fitting ellipse - circ.: circularity: 4pi(area/perim.^2) - feret: maximal feret diameter - intden: integrated density: mean*area - median: median grey - skew,kurt: skewness,kurtosis of the histogram of greys - %area: proportion of the image corresponding to the object - area_exc: area excluding holes - fractal: fractal dimension of the perimeter - skelarea: area of the one-pixel wide skeleton of the image - slope: slope of the cumulated histogram of greys - histcum1,2,3: grey level at quantiles 0.25, 0.5, 0.75 of the histogram of greys - nb1,2,3: number of objects after thresholding at the grey levels above - symetrieh,symetriev: index of horizontal,vertical symmetry - symetriehc,symetrievc: same but after thresholding at level histcum1 - convperim,convarea: perimeter,area of the convex hull of the object - fcons: contrast - thickr: thickness ratio: maximum thickness/mean thickness - esd: Equivalent Spherical Diameter - elongation: elongation index: major/minor - range: range of greys: max-min - centroids : sqrt(pow(xm-x,2)+ pow(ym-y,2)) - sr: index of variation of greys: 100*(stddev/range) - perimareaexc : perim/(sqrt(area_exc)) - feretareaexc : feret/(sqrt(area_exc)) - perimferet: index of the relative complexity of the perimeter: perim/feret - perimmajor: index of the relative complexity of the perimeter: perim/major - circex: (4*PI*area_exc)/(pow(perim,2)) - cdexc: (1/(sqrt(area_exc))) * sqrt(pow(xm-x,2)+pow(ym-y,2) features_skimage.csv.gz Table of morphological features recomputed with skimage.measure.regionprops on the ROIs produced by ZooProcess. See http://scikit-image.org/docs/dev/api/skimage.measure.html#skimage.measure.regionprops for documentation. inventory.tsv Tree view of the taxonomy and number of images in each taxon, displayed as text. With columns : - lineage_level1: taxonomic lineage corresponding to the level 1 classification - taxon_level1: name of the taxon corresponding to the level 1 classification - n: number of objects in each taxon class map.png Map of the sampling locations, to give an idea of the diversity sampled in this dataset. imgs Directory containing images of each object, named according to the object id objid and sorted in subdirectories according to their taxon.
-
Here, we provide plankton image data that was sorted with the web applications EcoTaxa and MorphoCluster. The data set was used for image classification tasks as described in Schröder et. al (in preparation) and does not include any geospatial or temporal meta-data. Plankton was imaged using the Underwater Vision Profiler 5 (Picheral et al. 2010) in various regions of the world's oceans between 2012-10-24 and 2017-08-08. This data publication consists of an archive containing "training.csv" (list of 392k training images for classification, validated using EcoTaxa), "validation.csv" (list of 196k validation images for classification, validated using EcoTaxa), "unlabeld.csv" (list of 1M unlabeled images), "morphocluster.csv" (1.2M objects validated using MorphoCluster, a subset of "unlabeled.csv" and "validation.csv") and the image files themselves. The CSV files each contain the columns "object_id" (a unique ID), "image_fn" (the relative filename), and "label" (the assigned name). The training and validation sets were sorted into 65 classes using the web application EcoTaxa (http://ecotaxa.obs-vlfr.fr). This data shows a severe class imbalance; the 10% most populated classes contain more than 80% of the objects and the class sizes span four orders of magnitude. The validation set and a set of additional 1M unlabeled images were sorted during the first trial of MorphoCluster (https://github.com/morphocluster). The images in this data set were sampled during RV Meteor cruises M92, M93, M96, M97, M98, M105, M106, M107, M108, M116, M119, M121, M130, M131, M135, M136, M137 and M138, during RV Maria S Merian cruises MSM22, MSM23, MSM40 and MSM49, during the RV Polarstern cruise PS88b and during the FLUXES1 experiment with RV Sarmiento de Gamboa. The following people have contributed to the sorting of the image data on EcoTaxa: Rainer Kiko, Tristan Biard, Benjamin Blanc, Svenja Christiansen, Justine Courboules, Charlotte Eich, Jannik Faustmann, Christine Gawinski, Augustin Lafond, Aakash Panchal, Marc Picheral, Akanksha Singh and Helena Hauss In Schröder et al. (in preparation), the training set serves as a source for knowledge transfer in the training of the feature extractor. The classification using MorphoCluster was conducted by Rainer Kiko. Used labels are operational and not yet matched to respective EcoTaxa classes.
-
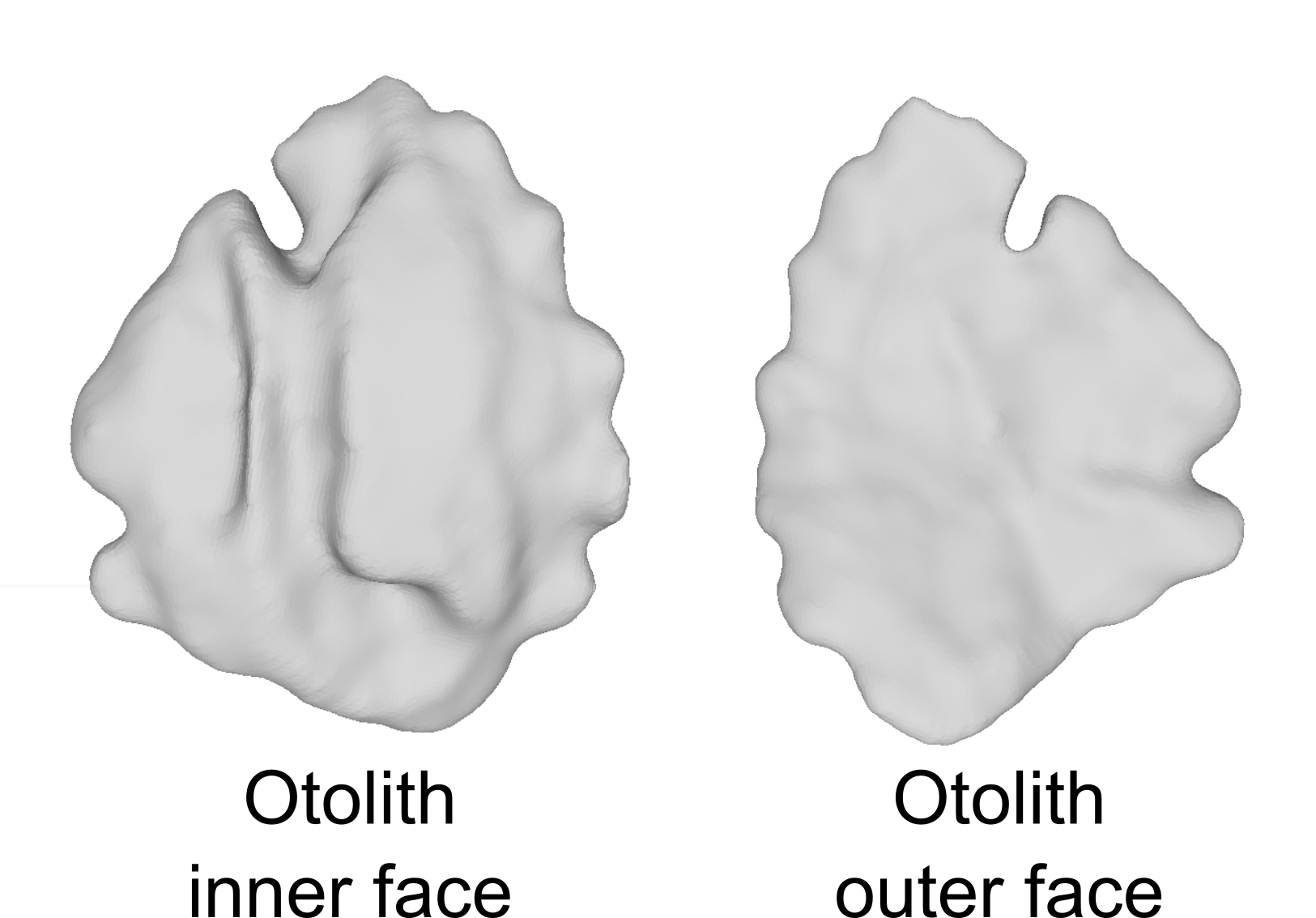
This dataset includes 3D sagittal left otolith meshes obtained from 339 individual red mullet (Mullus barbatus) specimens. These samples were collected from 17 distinct geographical locations spanning the whole Mediterranean Sea. Recorded biological parameters include fish total length (TL, ranging from 125 to 238 mm), total weight (W, ranging from 14.9 to 168.0 g), sex, and sexual maturity staging. The 3D otolith dataset consists of high-resolution meshes obtained through microtomography (29.2 m voxel size). The dataset provides valuable insights into the morphological variability and population structure of red mullet populations in the Mediterranean Sea.
-
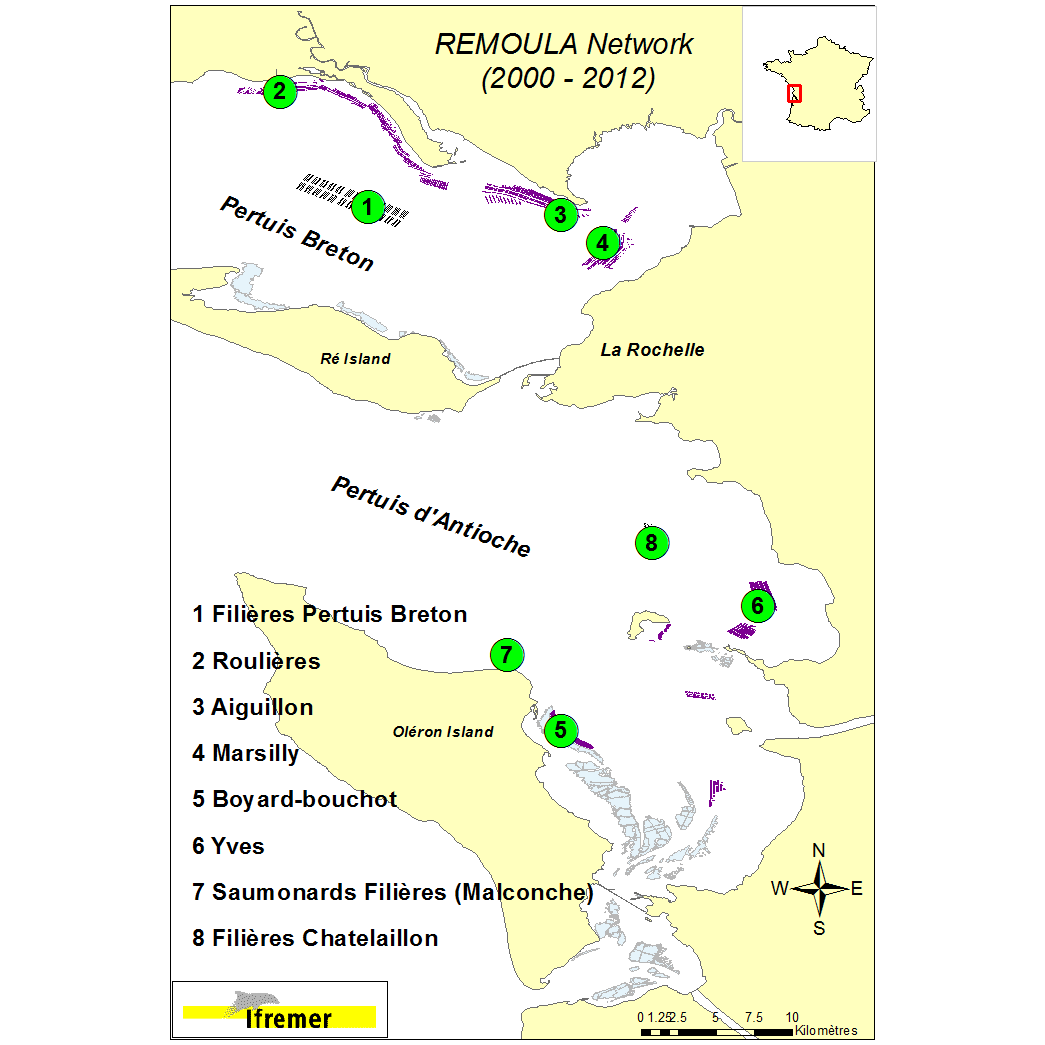
Concomitantly to the monitoring network of the Blue mussel growth in the Pertuis Charentais sounds, (available soon from SEANOE) high frequency temperature measurements were carried out on a regular basis and on all REMOULA monitoring stations. Temperature were recorded every 15’ on 7 experimental sites from 2010 to 2012. Two environemental conditions were tested, i.e. off shore and intertidal areas. The off-shore sites are located along the long lines mussel growout facilities (Filières Pertuis Breton, Saumonards Filières). Intertidally, temperature sensors were deployed on bouchot type mussel culture (wooden piles) (Roulières, Aiguillon, Marsilly, Boyard-bouchot, Yves). Due to the tidal cycle, the later are emersed on a regular basis – during this period of time, air temperature is recorded. The data set are presented in two ways: raw data (immersion-emersion values) and daily average (only immersed data). The daily average aims to represent the lasting period of mussel activity for further comparison with off shore conditions. For off-shore sites, daily averaged data are presented. For intertidal areas (bouchot type), the average is based upon the two daily high tides. Daily data are recorded in betwwen 2 hours before and after the high tide peak. Figures are presented per campaign and per site. Data temperature are recorded using Tidbit V2 logger (-39°C+75°C) and Sensor EN Optic STOWAWAY TEMP (-39+75°C) ONSET COMPUTER from 2000 to 2009 and NKE STPS30 probes (with and without chlorine system) and YSI 6600 from 2010 to 2012. Data storage is organized using the Quadrige data bank system. Coastal monitoring information are saved in the Coastal monitoring Quadrige information system.
-
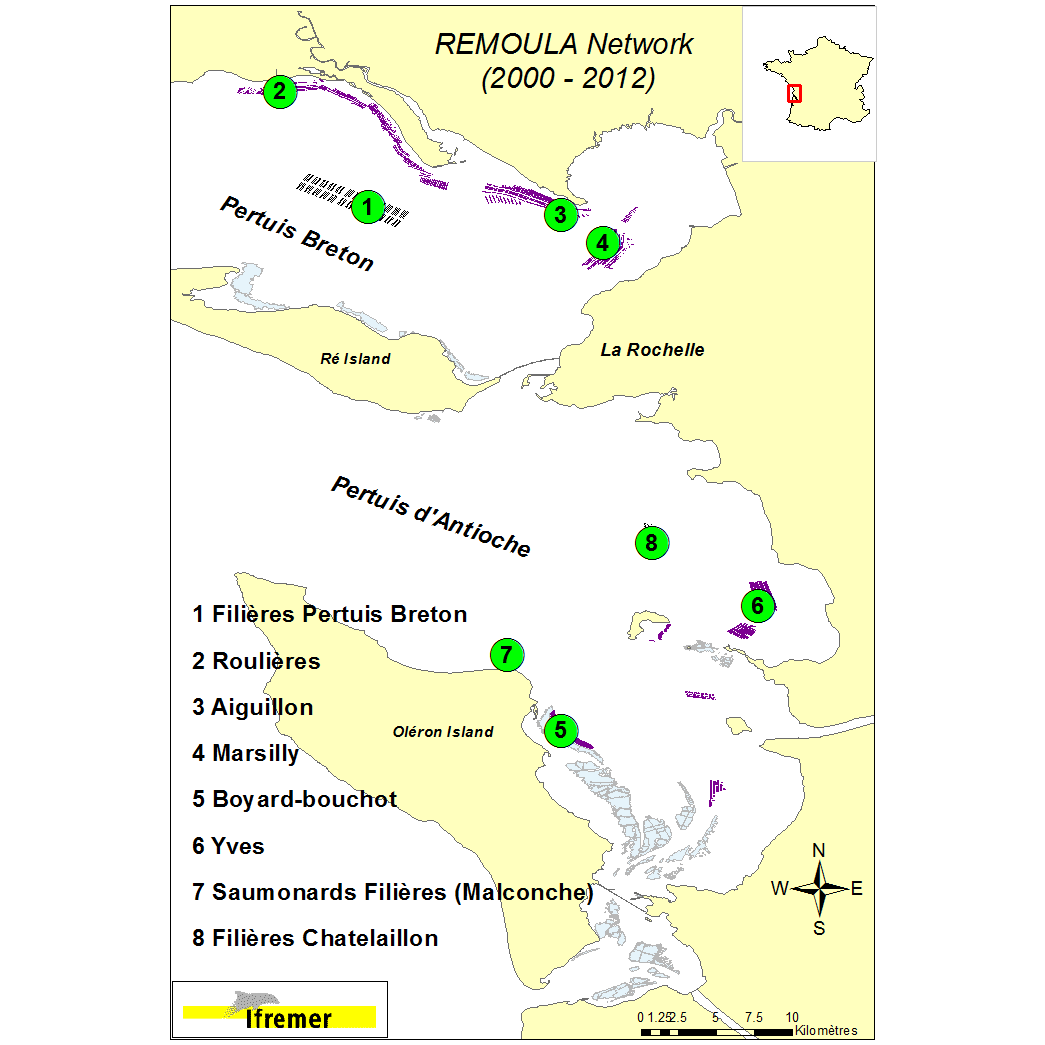
On a yearly basis, mussel farming produces around 13 000 metric tons of mussel in the Pertuis Charentais sounds (Pertuis Breton et Pertuis d’Antioche). This represents 23% of the Blue mussel (Mytilus edulis) production along the French Atlantic coastline. Production is characterized in this area by to types of growout systems: the ‘bouchot’ type culture using wooden poles(p) and off-shore longlines(l). Environmental conditions are mainly estuarine. Taking into account those cultural practices, the REMOULA monitoring network has been deployed since 2000 to provide baseline information to better understand the Blue mussel (Aiguillon(p) and Filières Pertuis Breton(l)) or trimester (Roulières(p), Marsilly(p), Yves(p), Boyard-bouchot(p) – completed by the stations Saumonards Filières(l) and Filières Chatellaillon 2(l) (2008 to 2012). Between 2000 and 2005 the mussel batches monitoring lasted 12 months whereas it lasted 15 months from 2006 to 2012. The initial batch of mussel is originating from the spat settlement the previous year and calibrated. On the 8 monitoring stations, calibrated mussel batches were deployed into bags (120 mussels/bag). Biometric data are measured during each monthly survey (30 individuals) and a mortality rate estimated. Additional information is obtained through biochemical analysis (proteins, lipids, carbohydrates) on 3 mussel pools of 10 individuals in 2000, 2001, 2002, 2003 and 2004. Moreover gametogenic data are collected on 15 mussels (2003 and 2004). The data storage is organized using the Quadrige2 system and characterized by individual field campaign. Coastal monitoring data Available parameters : Individual measurement : length, shell weight, dry meat weight. Width and height at the beginning and the campaign end. Average measurement : individual weight (3x10 individuals), Calculated data: Lawrence & Scott indicator, Walne and Mann indicator, meat indicator, mortality rate % Observations : Pinnotheres and Polydora infestations, parasites. Gametic status : identification of 8 maturation stages (0, 1, 2, 3A1, 3A2, 3B, 3C, 3D) according to Lubet (1959) and Suarez (2005) Biochemical data : analytical protocols - Proteins : Lowry et al (1951) modified Razet et al (1976). Lipids : Marsh and Weinstein (1966). Carbohydrates and Glycogen Dubois et al (1956). The shellfish biochemical proximate composition reviewed by Faury, Geairon, Moal, Pouvreau, Razet, Ropert and Soletchnik (2003).
-

Phenotypic plasticity, the ability of a single genotype to produce multiple phenotypes, is important for survival when species are faced with novel conditions. Theory predicts that range-edge populations will have greater phenotypic plasticity than core populations, but empirical examples from the wild are rare. The honeycomb worm, Sabellaria alveolata (L.), constructs the largest biogenic reefs in Europe, which support high biodiversity and numerous ecological functions. In order to assess the presence, causes and consequences of intraspecific variation in developmental plasticity and thermal adaptation in the honeycomb worm, we carried out common-garden experiments using the larvae of individuals sampled from along a latitudinal gradient covering the entire range of the species. We exposed larvae to three temperature treatments and measured phenotypic traits throughout development. We found phenotypic plasticity in larval growth rate but local adaptation in terms of larval period. The northern and southern range-edge populations of S. alveolata showed phenotypic plasticity for growth rate: growth rate increased as temperature treatment increased. In contrast, the core range populations showed no evidence of phenotypic plasticity. We present a rare case of range-edge plasticity at both the northern and southern range limit of species, likely caused by evolution of phenotypic plasticity during range expansion and its maintenance in highly heterogeneous environments. This dataset presents the raw image data collected for larval stages of Sabellaria alveolata from 5 populations across Europe and Northern Africa, exposed to 15, 20 and 25 C. Included are also opercular crown measurements used to estimate de size classes of individuals present in each population. All measurements made with the images collected are presented in an Excel spreadsheet, also available here.
-
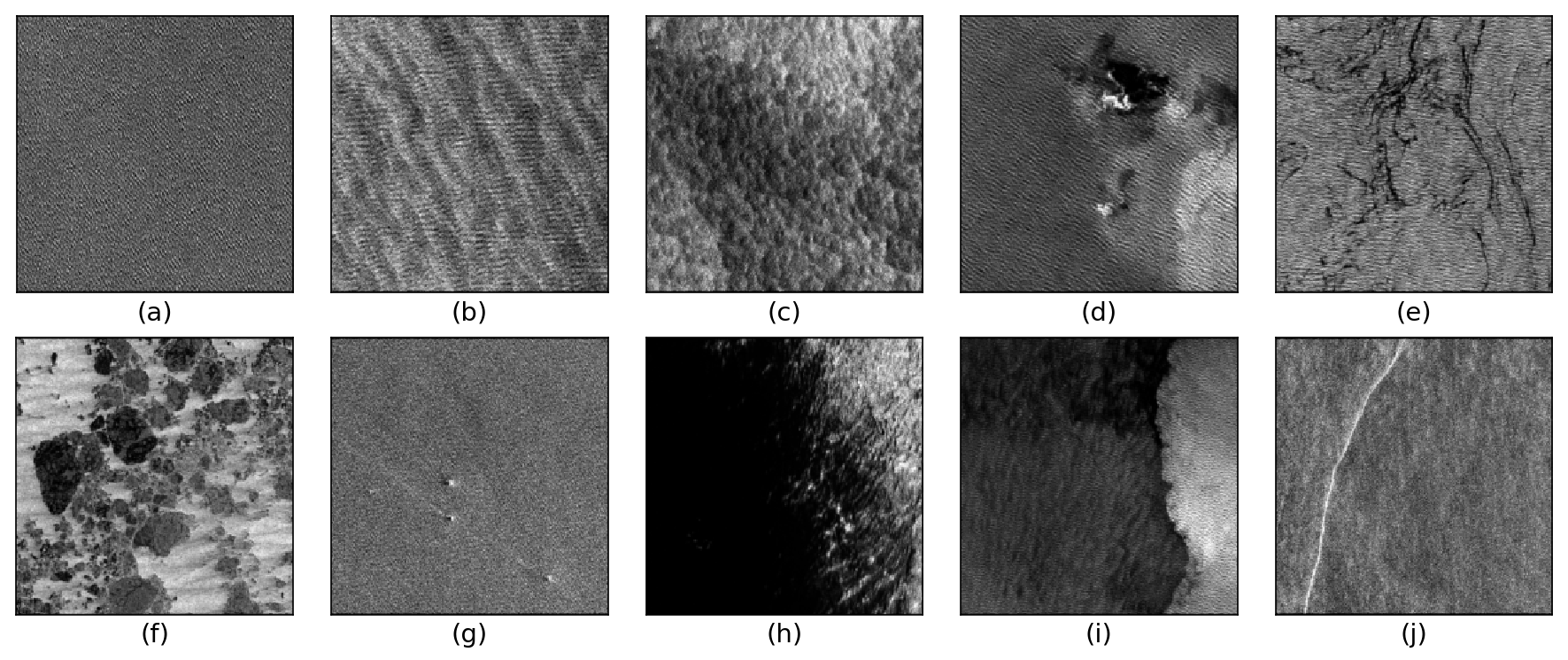
The TenGeoP-SARwv dataset is established based on the acquisitions of Sentinel-1A wave mode (WV) in VV polarization. This dataset consists of more than 37,000 SAR vignettes divided into ten defined geophysical categories, including both oceanic and meteorologic features. These images cover the entire open ocean and are manually selected from Sentinel-1A WV acquisitions in 2016. For each image, only one prevalent geophysical phenomena with its prescribed signature and texture is selected for labeling. The SAR images are processed into a quick-look image provided in the formats of PNG and GeoTIFF as well as the associated labels. They are convenient for both visual inspection and machine-learning-based methods exploitation. The proposed dataset is the first one involving different oceanic or atmospheric phenomena over the open ocean. It seeks to foster the development of strategies or approaches for massive ocean SAR image analysis. A key objective is to allow exploiting the full potential of Sentinel-1 WV SAR acquisitions, which are about 60,000 images per satellite per month and freely available. Such a dataset may be of value to a wide range of users and communities in deep learning, remote sensing, oceanography, and meteorology
-

The flat oyster Ostrea edulis is a European native species that once covered vast areas in the North Sea, on the Atlantic coast and in other European coastal waters including the Mediterranean region. All these populations have been heavily fished by dredging over the last three centuries. More recently, the emergence of parasites combined with the proliferation of various predators and many human-induced additional stressors have caused a dramatic decrease in the last remaining flat oyster populations. Today, this species has disappeared from many locations in Europe and is registered on the OSPAR (Oslo-Paris Convention for the Protection of the Marine environment of the North-East Atlantic) list of threatened and/or declining species (see https://www.ospar.org/work-areas/bdc/species-habitats/list-of-threatened-declining-species-habitats). In that context, since 2018, the Flat Oyster REcoVERy project (FOREVER) has been promoting the reestablishment of native oysters in Brittany (France). This multi-partner project, involving the CRC (Comité Régional de la Conchyliculture), IFREMER (Institut Français de Recherche pour l’Exploitation de la Mer), ESITC (École Supérieure d’Ingénieurs des Travaux de la Construction) Caen and Cochet Environnement, has consisted of (1) inventorying and evaluating the status of the main wild flat oyster populations across Brittany, (2) making detailed analysis of the two largest oyster beds in the bays of Brest and Quiberon to improve understanding of flat oyster ecology and recruitment variability and to suggest possible ways of improving recruitment, and (3) proposing practical measures for the management of wild beds in partnership with members of the shellfish industry and marine managers. the final report of this project is available on Archimer : https://doi.org/10.13155/79506. This survey is part of the task 1 of the FOREVER, which took place between 2017-2021. Some previous data, acquired with the same methodology and within the same geographic area have been also added to this dataset. These data were collected during 30 intertidal and diving surveys in various bays and inlets of the coast of Bretagne. The localization of these surveys has been guided by the help of historical maps. In the field, the methodology was simple enough to be easily implemented regardless of the configuration of the sampled site. The intertidal survey was conducted at very low tide (tidal range > 100) to sample the 0-1m level. Sampling was carried out randomly or systematically following the low water line. Where possible (in terms of visibility and accessibility), dive surveys were also carried out (0-10m depth), along 100m transects, using the same methodology of counting in a 1m2 quadrat. As often as possible, geo-referenced photographs were taken to show the appearance, density and habitat where Ostrea edulis was present. All these pictures are available in the image bank file. Overall, this dataset contains a total of 300 georeferenced records, where flat oysters have been observed. The dataset file contains also information concerning the surrounding habitat description and is organized according the OSPAR recommendations. This publication gives also a map, under a kml format showing each occurrence and its characteristics. This work was done in the framework of the following research project: " Inventaire, diagnostic écologique et restauration des principaux bancs d’huitres plates en Bretagne : le projet FOREVER. Contrat FEAMP 17/2215675".
-
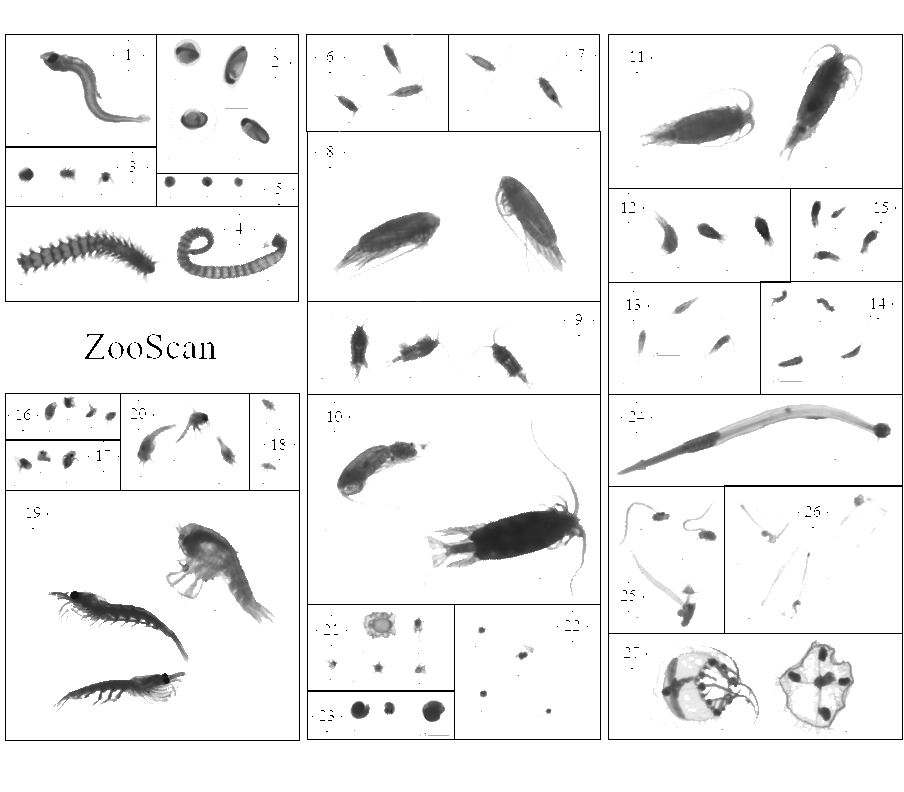
This dataset is composed of 1,153,507 zooplankton individuals, zooplankton parts, non-living particles and imaging artefacts, ranging from 300 µm to 3.39 mm Equivalent Spherical Diameter, individually imaged and measured with the ZooScan (Gorsky et al., 2010). The objects were sorted in 127 taxonomic and morphological groups. The imaged objects originate from samples collected on the Bay of Biscay continental shelf, in spring, from 2004 to 2016 during the PELGAS ecosystemic surveys (Doray et al., 2018). The samples were collected with a WP2 200 µm mesh size fitted with a Hydrobios (back-run stop) mechanical flowmeter, generally from 100 m depth to the surface, or 5 m above the sea floor (if bottom depth less than 100 m) in vertical hauls, at night. From 2004 to 2006, vertical WP2 net tows were performed in the anchovy core distribution area in the southern Bay of Biscay and North of it until the Loire estuary only. Since 2009, WP2 sampling has been carried out at all PELGAS stations, up to the southern coast of Brittany. The samples were preserved in 4% buffered formaldehyde seawater solution directly after collection, until 2019-2020 where they were imaged with the ZooScan, in the lab, on land. Each imaged object is geolocated, associated to a station, a cruise, a year and other metadata that enable the reconstruction of quantitative zooplankton communities for ecological studies (i.e. Grandrémy et al., 2023a). Each object is described by 46 morphological and grey level based features (8 bits encoding, 0 = black, 255 = white), including size, automatically extracted on each individual image by the Zooprocess. Each object was taxonomically identified using the web based application Ecotaxa with built-in, random forest and CNN based, semi-automatic sorting tools followed by expert validation or correction (Picheral et al., 2017). This dataset is intended to be used for ecological studies as well as machine learning applied to plankton studies. The archive contains: - One tab separated file (PELGAS ZooScan zooplankton dataset) containing all data and metadata associated to each imaged and identified object. Metadata and features are in columns (n =71) and objects are in rows (n = 1,153,507). - One comma separated file containing the name, type, definition and unit of each field (column) in the .tsv (dataset_descriptor_zooscan). - One comma separated file containing the taxonomic list of the dataset, with counts and nature of the content of the category, i.e. “T” for taxonomical category, and “M” for morphological category (taxonomy_descriptor_zooscan). - A individual_images directory containing images of each imaged object sorted in subdirectories named according to objects’ identifications object_taxon appended to an Ecotaxa internal taxon numerical id classif_id (i.e. taxon__123456789) across years and sampling stations. Within subdirectories, each object is named after its unique internal Ecotaxa identifier, objid. - A Map of the sampling station location over the 2004-2016 period
-
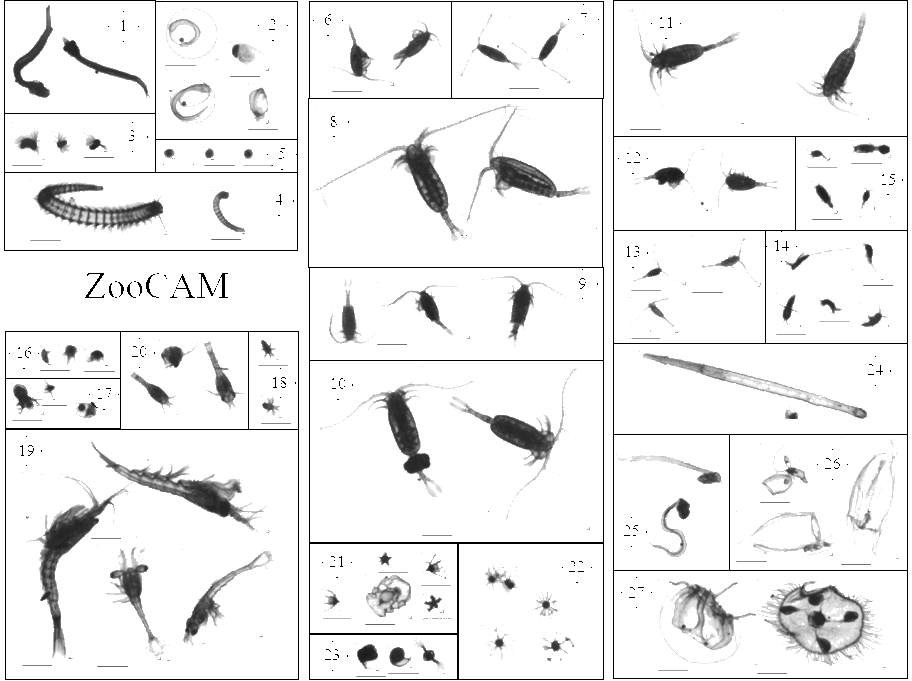
This dataset is composed of 702,111 zooplankton individuals, zooplankton pieces, non-living particles and imaging artefacts, ranging from 300 µm to 3.39 mm Equivalent Spherical Diameter, individually imaged and measured with the ZooCAM (Colas et al., 2018). The objects were sorted in 127 taxonomic and morphological groups. The imaged objects originate from samples collected on the Bay of Biscay continental shelf, in spring, from 2016 to 2019 during the PELGAS ecosystemic surveys (Doray et al., 2018). The samples were collected with a WP2 200 µm mesh size fitted with a Hydrobios (back-run stop) mechanical flowmeter, generally from 100 m depth to the surface, or 5 m above the sea floor (if bottom depth less than 100 m) in vertical hauls, at night. The samples were imaged on board, live, after collection and subsampling, and preserved in 4% buffered formaldehyde seawater. Each imaged object is geolocated, associated to a station, a cruise, a year and other metadata that enable the reconstruction of quantitative zooplankton communities for ecological studies (i.e. Grandrémy et al., 2023a). Each object is described by 52 morphological and grey level based features (8 bits encoding, 0 = black, 255 = white), including size, automatically extracted on each individual image by the ZooCAM software. Each object was taxonomically identified using the ZooCAM software and the web based application Ecotaxa with built-in, random forest and CNN based, semi-automatic sorting tools followed by expert validation or correction (Picheral et al., 2017). Images from 2016-2017 contain ROI bounding box limits, metadata at the bottom of each image, and non-homogenised background within and around the ROI bounding box; Images from 2018 contain non-homogenised background within the ROI bounding box only; images from 2019 have a completely homogeneous and thresholded background around the object. The differences arose from successive ZooCAM software updates that do not modify the calculation of object’s features. This dataset is intended to be used for ecological studies as well as machine learning applied to plankton studies. The archive contains : - One tab separated file (PELGAS ZooCAM zooplankton dataset) containing all data and metadata associated to each imaged and identified object. Metadata and features are in columns (n =72) and objects are in rows (n = 702,111). - One comma separated file containing the name, type, definition and unit of each field (column) in the .tsv (dataset descriptor zoocam). - One comma separated file containing the taxonomic list of the dataset, with counts and nature of the content of the category, i.e. “T” for taxonomical category, and “M” for morphological category (taxonomy descriptor zoocam). - A individual_images directory containing images of each object, named according to the object id objid and sorted in subdirectories according to their taxonomic identification, across years and sampling stations. - A Map of the sampling station location over the 2016-2019 period.
 Catalogue PIGMA
Catalogue PIGMA